Is There a Relationship between DNA Methylation and Phenotypic Plasticity in Invertebrates?
- PMID: 22232607
- PMCID: PMC3249382
- DOI: 10.3389/fphys.2011.00116
Is There a Relationship between DNA Methylation and Phenotypic Plasticity in Invertebrates?
Abstract
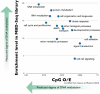
There is a significant amount of variation in DNA methylation characteristics across organisms. Likewise, the biological role of DNA methylation varies across taxonomic lineages. The complexity of DNA methylation patterns in invertebrates has only recently begun to be characterized in-depth. In some invertebrate species that have been examined to date, methylated DNA is found primarily within coding regions and patterning is closely associated with gene function. Here we provide a perspective on the potential role of DNA methylation in these invertebrates with a focus on how limited methylation may contribute to increased phenotypic plasticity in highly fluctuating environments. Specifically, limited methylation could facilitate a variety of transcriptional opportunities including access to alternative transcription start sites, increasing sequence mutations, exon skipping, and transient methylation.
Keywords: adaptation; epigenetic; methylation; oyster; plasticity.
Figures

Similar articles
-
Epigenetic features in the oyster Crassostrea gigas suggestive of functionally relevant promoter DNA methylation in invertebrates.Front Physiol. 2014 Apr 7;5:129. doi: 10.3389/fphys.2014.00129. eCollection 2014. Front Physiol. 2014. PMID: 24778620 Free PMC article.
-
Patterns of DNA methylation in animals: an ecotoxicological perspective.Integr Comp Biol. 2014 Jul;54(1):77-86. doi: 10.1093/icb/icu025. Epub 2014 Apr 29. Integr Comp Biol. 2014. PMID: 24785828 Review.
-
Evolutionary Transition of Promoter and Gene Body DNA Methylation across Invertebrate-Vertebrate Boundary.Mol Biol Evol. 2016 Apr;33(4):1019-28. doi: 10.1093/molbev/msv345. Epub 2015 Dec 29. Mol Biol Evol. 2016. PMID: 26715626 Free PMC article.
-
Genome-Wide DNA Methylation Signatures of Sea Cucumber Apostichopus japonicus during Environmental Induced Aestivation.Genes (Basel). 2020 Aug 31;11(9):1020. doi: 10.3390/genes11091020. Genes (Basel). 2020. PMID: 32877994 Free PMC article.
-
Ecotoxicological epigenetics in invertebrates: Emerging tool for the evaluation of present and past pollution burden.Chemosphere. 2021 Nov;282:131026. doi: 10.1016/j.chemosphere.2021.131026. Epub 2021 Jun 2. Chemosphere. 2021. PMID: 34111635 Review.
Cited by
-
Inheritance and Variation of Genomic DNA Methylation in Diploid and Triploid Pacific Oyster (Crassostrea gigas).Mar Biotechnol (NY). 2016 Feb;18(1):124-32. doi: 10.1007/s10126-015-9674-4. Epub 2015 Nov 19. Mar Biotechnol (NY). 2016. PMID: 26585587
-
Cross-species analysis of genic GC3 content and DNA methylation patterns.Genome Biol Evol. 2013;5(8):1443-56. doi: 10.1093/gbe/evt103. Genome Biol Evol. 2013. PMID: 23833164 Free PMC article.
-
No more non-model species: the promise of next generation sequencing for comparative immunology.Dev Comp Immunol. 2014 Jul;45(1):56-66. doi: 10.1016/j.dci.2014.01.022. Epub 2014 Feb 6. Dev Comp Immunol. 2014. PMID: 24508980 Free PMC article. Review.
-
Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies.Proc Natl Acad Sci U S A. 2015 Nov 10;112(45):13970-5. doi: 10.1073/pnas.1515937112. Epub 2015 Oct 19. Proc Natl Acad Sci U S A. 2015. PMID: 26483466 Free PMC article.
-
Insecticide exposure affects intergenerational patterns of DNA methylation in the Colorado potato beetle, Leptinotarsa decemlineata.Evol Appl. 2020 Nov 25;14(3):746-757. doi: 10.1111/eva.13153. eCollection 2021 Mar. Evol Appl. 2020. PMID: 33767749 Free PMC article.
References
LinkOut - more resources
Full Text Sources

