RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity
- PMID: 22223809
- PMCID: PMC3295403
- DOI: 10.1093/jxb/err434
RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity
Abstract
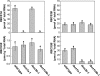
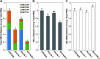
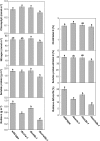
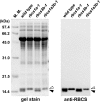
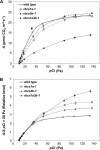
Ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit (RBCS) is encoded by a nuclear RBCS multigene family in many plant species. The contribution of the RBCS multigenes to accumulation of Rubisco holoenzyme and photosynthetic characteristics remains unclear. T-DNA insertion mutants of RBCS1A (rbcs1a-1) and RBCS3B (rbcs3b-1) were isolated among the four Arabidopsis RBCS genes, and a double mutant (rbcs1a3b-1) was generated. RBCS1A mRNA was not detected in rbcs1a-1 and rbcs1a3b-1, while the RBCS3B mRNA level was suppressed to ∼20% of the wild-type level in rbcs3b-1 and rbcs1a3b-1 leaves. As a result, total RBCS mRNA levels declined to 52, 79, and 23% of the wild-type level in rbcs1a-1, rbcs3b-1, and rbcs1a3b-1, respectively. Rubisco contents showed declines similar to total RBCS mRNA levels, and the ratio of Rubisco-nitrogen to total nitrogen was 62, 78, and 40% of the wild-type level in rbcs1a-1, rbcs3b-1, and rbcs1a3b-1, respectively. The effects of RBCS1A and RBCS3B mutations in rbcs1a3b-1 were clearly additive. The rates of CO(2) assimilation at ambient CO(2) of 40 Pa were reduced with decreased Rubisco contents in the respective mutant leaves. Although the RBCS composition in the Rubisco holoenzyme changed, the CO(2) assimilation rates per unit of Rubisco content were the same irrespective of the genotype. These results clearly indicate that RBCS1A and RBCS3B contribute to accumulation of Rubisco in Arabidopsis leaves and that these genes work additively to yield sufficient Rubisco for photosynthetic capacity. It is also suggested that the RBCS composition in the Rubisco holoenzyme does not affect photosynthesis under the present ambient [CO(2)] conditions.
Figures
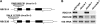

Similar articles
-
Effect of individual suppression of RBCS multigene family on Rubisco contents in rice leaves.Plant Cell Environ. 2012 Mar;35(3):546-53. doi: 10.1111/j.1365-3040.2011.02434.x. Epub 2011 Oct 24. Plant Cell Environ. 2012. PMID: 21951138
-
Availability of Rubisco small subunit up-regulates the transcript levels of large subunit for stoichiometric assembly of its holoenzyme in rice.Plant Physiol. 2012 Sep;160(1):533-40. doi: 10.1104/pp.112.201459. Epub 2012 Jul 17. Plant Physiol. 2012. PMID: 22811433 Free PMC article.
-
Structure of Rubisco from Arabidopsis thaliana in complex with 2-carboxyarabinitol-1,5-bisphosphate.Acta Crystallogr D Struct Biol. 2018 Jan 1;74(Pt 1):1-9. doi: 10.1107/S2059798317017132. Epub 2018 Jan 1. Acta Crystallogr D Struct Biol. 2018. PMID: 29372894 Free PMC article.
-
Photosynthesis, plant growth and N allocation in transgenic rice plants with decreased Rubisco under CO2 enrichment.J Exp Bot. 2000 Feb;51 Spec No:383-9. doi: 10.1093/jexbot/51.suppl_1.383. J Exp Bot. 2000. PMID: 10938846 Review.
-
Rubisco gene expression in C4 plants.J Exp Bot. 2008;59(7):1625-34. doi: 10.1093/jxb/erm368. Epub 2008 Mar 5. J Exp Bot. 2008. PMID: 18325924 Review.
Cited by
-
Transcriptome Analysis of the Effects of Grafting Interstocks on Apple Rootstocks and Scions.Int J Mol Sci. 2023 Jan 2;24(1):807. doi: 10.3390/ijms24010807. Int J Mol Sci. 2023. PMID: 36614250 Free PMC article.
-
Ascophyllum nodosum Seaweed Extract Alleviates Drought Stress in Arabidopsis by Affecting Photosynthetic Performance and Related Gene Expression.Front Plant Sci. 2017 Aug 3;8:1362. doi: 10.3389/fpls.2017.01362. eCollection 2017. Front Plant Sci. 2017. PMID: 28824691 Free PMC article.
-
Disclosing proteins in the leaves of cork oak plants associated with the immune response to Phytophthora cinnamomi inoculation in the roots: A long-term proteomics approach.PLoS One. 2021 Jan 22;16(1):e0245148. doi: 10.1371/journal.pone.0245148. eCollection 2021. PLoS One. 2021. PMID: 33481834 Free PMC article.
-
Identification of source-sink tissues in the leaf of Chinese cabbage (Brassica rapa ssp. pekinensis) by carbohydrate content and transcriptomic analysis.Genes Genomics. 2020 Jan;42(1):13-24. doi: 10.1007/s13258-019-00873-z. Epub 2019 Oct 14. Genes Genomics. 2020. PMID: 31612374
-
Arabidopsis RabF1 (ARA6) Is Involved in Salt Stress and Dark-Induced Senescence (DIS).Int J Mol Sci. 2017 Feb 1;18(2):309. doi: 10.3390/ijms18020309. Int J Mol Sci. 2017. PMID: 28157156 Free PMC article.
References
-
- Andersson I. Large structures at high resolution: the 1.6 angstrom crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate. Journal of Molecular Biology. 1996;259:160–174. - PubMed
-
- Dean C, Pichersky E, Dunsmuir P. Structure, evolution, and regulation of rbcS genes in higher plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1989;40:415–439.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

