Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the Lamina joint of rice
- PMID: 22207574
- PMCID: PMC3269869
- DOI: 10.1105/tpc.111.093419
Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the Lamina joint of rice
Abstract
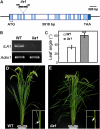
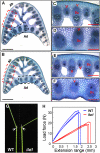
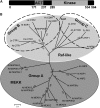
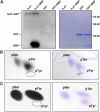
Mitogen-activated protein kinase kinase kinases (MAPKKKs), which function at the top level of mitogen-activated protein kinase cascades, are clustered into three groups. However, no Group C Raf-like MAPKKKs have yet been functionally identified. We report here the characterization of a rice (Oryza sativa) mutant, increased leaf angle1 (ila1), resulting from a T-DNA insertion in a Group C MAPKKK gene. The increased leaf angle in ila1 is caused by abnormal vascular bundle formation and cell wall composition in the leaf lamina joint, as distinct from the mechanism observed in brassinosteroid-related mutants. Phosphorylation assays revealed that ILA1 is a functional kinase with Ser/Thr kinase activity. ILA1 is predominantly resident in the nucleus and expressed in the vascular bundles of leaf lamina joints. Yeast two-hybrid screening identified six closely related ILA1 interacting proteins (IIPs) of unknown function. Using representative IIPs, the interaction of ILA1 and IIPs was confirmed in vivo. IIPs were localized in the nucleus and showed transactivation activity. Furthermore, ILA1 could phosphorylate IIP4, indicating that IIPs may be the downstream substrates of ILA1. Microarray analyses of leaf lamina joints provided additional evidence for alterations in mechanical strength in ila1. ILA1 is thus a key factor regulating mechanical tissue formation at the leaf lamina joint.
Figures
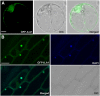
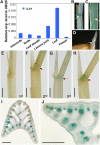

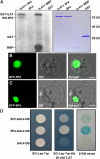
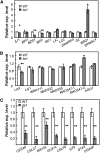
Similar articles
-
AUXIN RESPONSE FACTORS 6 and 17 control the flag leaf angle in rice by regulating secondary cell wall biosynthesis of lamina joints.Plant Cell. 2021 Sep 24;33(9):3120-3133. doi: 10.1093/plcell/koab175. Plant Cell. 2021. PMID: 34245297 Free PMC article.
-
An Uncanonical CCCH-Tandem Zinc-Finger Protein Represses Secondary Wall Synthesis and Controls Mechanical Strength in Rice.Mol Plant. 2018 Jan 8;11(1):163-174. doi: 10.1016/j.molp.2017.11.004. Epub 2017 Nov 22. Mol Plant. 2018. PMID: 29175437
-
A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice.Plant Physiol. 2010 Feb;152(2):876-90. doi: 10.1104/pp.109.149856. Epub 2009 Dec 9. Plant Physiol. 2010. PMID: 20007444 Free PMC article.
-
Identification and functional analysis of mitogen-activated protein kinase kinase kinase (MAPKKK) genes in canola (Brassica napus L.).J Exp Bot. 2014 May;65(8):2171-88. doi: 10.1093/jxb/eru092. Epub 2014 Mar 6. J Exp Bot. 2014. PMID: 24604738 Free PMC article.
-
Leaf direction: Lamina joint development and environmental responses.Plant Cell Environ. 2021 Aug;44(8):2441-2454. doi: 10.1111/pce.14065. Epub 2021 May 20. Plant Cell Environ. 2021. PMID: 33866581 Review.
Cited by
-
SMALL PLANT AND ORGAN 1 (SPO1) Encoding a Cellulose Synthase-like Protein D4 (OsCSLD4) Is an Important Regulator for Plant Architecture and Organ Size in Rice.Int J Mol Sci. 2023 Nov 30;24(23):16974. doi: 10.3390/ijms242316974. Int J Mol Sci. 2023. PMID: 38069299 Free PMC article.
-
WGCNA analysis of the effect of exogenous BR on leaf angle of maize mutant lpa1.Sci Rep. 2024 Mar 4;14(1):5238. doi: 10.1038/s41598-024-55835-7. Sci Rep. 2024. PMID: 38433245 Free PMC article.
-
The Rice Basic Helix-Loop-Helix 79 (OsbHLH079) Determines Leaf Angle and Grain Shape.Int J Mol Sci. 2020 Mar 18;21(6):2090. doi: 10.3390/ijms21062090. Int J Mol Sci. 2020. PMID: 32197452 Free PMC article.
-
OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.).BMC Plant Biol. 2014 Jun 6;14:158. doi: 10.1186/1471-2229-14-158. BMC Plant Biol. 2014. PMID: 24906444 Free PMC article.
-
An Analysis of Natural Variation Reveals That OsFLA2 Controls Flag Leaf Angle in Rice (Oryza sativa L.).Front Plant Sci. 2022 Jun 23;13:906912. doi: 10.3389/fpls.2022.906912. eCollection 2022. Front Plant Sci. 2022. PMID: 35812967 Free PMC article.
References
-
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 - PubMed
-
- Bergmann D.C., Lukowitz W., Somerville C.R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304: 1494–1497 - PubMed
-
- Cao H., Chen S. (1995). Brassinosteroid-induced rice lamina joint inclination and its relation to indole-3-acetic acid and ethylene. Plant Growth Regul. 16: 189–196
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

