Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing
- PMID: 22197930
- PMCID: PMC3267896
- DOI: 10.1038/ng.1041
Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing
Abstract
We sequenced eight melanoma exomes to identify new somatic mutations in metastatic melanoma. Focusing on the mitogen-activated protein (MAP) kinase kinase kinase (MAP3K) family, we found that 24% of melanoma cell lines have mutations in the protein-coding regions of either MAP3K5 or MAP3K9. Structural modeling predicted that mutations in the kinase domain may affect the activity and regulation of these protein kinases. The position of the mutations and the loss of heterozygosity of MAP3K5 and MAP3K9 in 85% and 67% of melanoma samples, respectively, together suggest that the mutations are likely to be inactivating. In in vitro kinase assays, MAP3K5 I780F and MAP3K9 W333* variants had reduced kinase activity. Overexpression of MAP3K5 or MAP3K9 mutants in HEK293T cells reduced the phosphorylation of downstream MAP kinases. Attenuation of MAP3K9 function in melanoma cells using siRNA led to increased cell viability after temozolomide treatment, suggesting that decreased MAP3K pathway activity can lead to chemoresistance in melanoma.
Figures



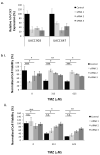
Similar articles
-
Somatic mutations in MAP3K5 attenuate its proapoptotic function in melanoma through increased binding to thioredoxin.J Invest Dermatol. 2014 Feb;134(2):452-460. doi: 10.1038/jid.2013.365. Epub 2013 Sep 5. J Invest Dermatol. 2014. PMID: 24008424 Free PMC article.
-
Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma.Nat Genet. 2011 Dec 25;44(2):133-9. doi: 10.1038/ng.1026. Nat Genet. 2011. PMID: 22197931
-
Molecular determinants of melanoma malignancy: selecting targets for improved efficacy of chemotherapy.Mol Cancer Ther. 2009 Mar;8(3):636-47. doi: 10.1158/1535-7163.MCT-08-0749. Epub 2009 Mar 10. Mol Cancer Ther. 2009. PMID: 19276165 Free PMC article.
-
Role of the MEK inhibitor trametinib in the treatment of metastatic melanoma.Future Oncol. 2014;10(9):1559-70. doi: 10.2217/fon.14.89. Future Oncol. 2014. PMID: 25145427 Review.
-
Melanoma Treatments: Advances and Mechanisms.J Cell Physiol. 2015 Nov;230(11):2626-33. doi: 10.1002/jcp.25019. J Cell Physiol. 2015. PMID: 25899612 Review.
Cited by
-
Advances in personalized targeted treatment of metastatic melanoma and non-invasive tumor monitoring.Front Oncol. 2013 Mar 19;3:54. doi: 10.3389/fonc.2013.00054. eCollection 2013. Front Oncol. 2013. PMID: 23515890 Free PMC article.
-
The role of altered nucleotide excision repair and UVB-induced DNA damage in melanomagenesis.Int J Mol Sci. 2013 Jan 9;14(1):1132-51. doi: 10.3390/ijms14011132. Int J Mol Sci. 2013. PMID: 23303275 Free PMC article. Review.
-
Melanoma: from mutations to medicine.Genes Dev. 2012 Jun 1;26(11):1131-55. doi: 10.1101/gad.191999.112. Genes Dev. 2012. PMID: 22661227 Free PMC article. Review.
-
Immunotherapy in melanoma: advances, pitfalls, and future perspectives.Front Mol Biosci. 2024 Jun 28;11:1403021. doi: 10.3389/fmolb.2024.1403021. eCollection 2024. Front Mol Biosci. 2024. PMID: 39086722 Free PMC article. Review.
-
Promoter methylation-regulated miR-148a-3p inhibits lung adenocarcinoma (LUAD) progression by targeting MAP3K9.Acta Pharmacol Sin. 2022 Nov;43(11):2946-2955. doi: 10.1038/s41401-022-00893-8. Epub 2022 Apr 6. Acta Pharmacol Sin. 2022. PMID: 35388129 Free PMC article.
References
-
-
Materials and Methods are available as supporting material on Nature Genetics Online.
-
-
- Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

