SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage
- PMID: 22174690
- PMCID: PMC3234236
- DOI: 10.1371/journal.ppat.1002433
SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage
Abstract
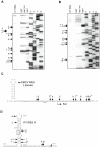
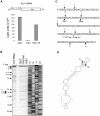
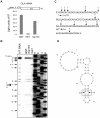
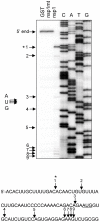
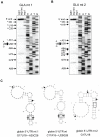
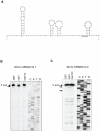
SARS coronavirus (SCoV) nonstructural protein (nsp) 1, a potent inhibitor of host gene expression, possesses a unique mode of action: it binds to 40S ribosomes to inactivate their translation functions and induces host mRNA degradation. Our previous study demonstrated that nsp1 induces RNA modification near the 5'-end of a reporter mRNA having a short 5' untranslated region and RNA cleavage in the encephalomyocarditis virus internal ribosome entry site (IRES) region of a dicistronic RNA template, but not in those IRES elements from hepatitis C or cricket paralysis viruses. By using primarily cell-free, in vitro translation systems, the present study revealed that the nsp1 induced endonucleolytic RNA cleavage mainly near the 5' untranslated region of capped mRNA templates. Experiments using dicistronic mRNAs carrying different IRESes showed that nsp1 induced endonucleolytic RNA cleavage within the ribosome loading region of type I and type II picornavirus IRES elements, but not that of classical swine fever virus IRES, which is characterized as a hepatitis C virus-like IRES. The nsp1-induced RNA cleavage of template mRNAs exhibited no apparent preference for a specific nucleotide sequence at the RNA cleavage sites. Remarkably, SCoV mRNAs, which have a 5' cap structure and 3' poly A tail like those of typical host mRNAs, were not susceptible to nsp1-mediated RNA cleavage and importantly, the presence of the 5'-end leader sequence protected the SCoV mRNAs from nsp1-induced endonucleolytic RNA cleavage. The escape of viral mRNAs from nsp1-induced RNA cleavage may be an important strategy by which the virus circumvents the action of nsp1 leading to the efficient accumulation of viral mRNAs and viral proteins during infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
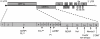




Similar articles
-
Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA.J Virol. 2012 Oct;86(20):11128-37. doi: 10.1128/JVI.01700-12. Epub 2012 Aug 1. J Virol. 2012. PMID: 22855488 Free PMC article.
-
Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation.J Virol. 2012 Dec;86(24):13598-608. doi: 10.1128/JVI.01958-12. Epub 2012 Oct 3. J Virol. 2012. PMID: 23035226 Free PMC article.
-
A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein.Nat Struct Mol Biol. 2009 Nov;16(11):1134-40. doi: 10.1038/nsmb.1680. Epub 2009 Oct 18. Nat Struct Mol Biol. 2009. PMID: 19838190 Free PMC article.
-
Mechanisms of Coronavirus Nsp1-Mediated Control of Host and Viral Gene Expression.Cells. 2021 Feb 2;10(2):300. doi: 10.3390/cells10020300. Cells. 2021. PMID: 33540583 Free PMC article. Review.
-
I(nsp1)ecting SARS-CoV-2-ribosome interactions.Commun Biol. 2021 Jun 10;4(1):715. doi: 10.1038/s42003-021-02265-0. Commun Biol. 2021. PMID: 34112887 Free PMC article. Review.
Cited by
-
Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA.J Virol. 2012 Oct;86(20):11128-37. doi: 10.1128/JVI.01700-12. Epub 2012 Aug 1. J Virol. 2012. PMID: 22855488 Free PMC article.
-
A common strategy for host RNA degradation by divergent viruses.J Virol. 2012 Sep;86(17):9527-30. doi: 10.1128/JVI.01230-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740404 Free PMC article.
-
Screening of FDA-approved compound library identifies potential small-molecule inhibitors of SARS-CoV-2 non-structural proteins NSP1, NSP4, NSP6 and NSP13: molecular modeling and molecular dynamics studies.J Proteins Proteom. 2021;12(3):161-175. doi: 10.1007/s42485-021-00067-w. Epub 2021 Jun 9. J Proteins Proteom. 2021. PMID: 34121824 Free PMC article.
-
The pandemic COVID-19: a tale of viremia, cellular oxidation and immune dysfunction.Pan Afr Med J. 2020 Jul 15;36:188. doi: 10.11604/pamj.2020.36.188.23476. eCollection 2020. Pan Afr Med J. 2020. PMID: 32952832 Free PMC article.
-
Review Devil's tools: SARS-CoV-2 antagonists against innate immunity.Curr Res Virol Sci. 2021;2:100013. doi: 10.1016/j.crviro.2021.100013. Epub 2021 Nov 18. Curr Res Virol Sci. 2021. PMID: 34812428 Free PMC article. Review.
References
-
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
-
- Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, et al. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. - PubMed
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. - PubMed
-
- Li W, Shi Z, Yu M, Ren W, Smith C, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

