Bioinformatic analysis and post-translational modification crosstalk prediction of lysine acetylation
- PMID: 22164248
- PMCID: PMC3229533
- DOI: 10.1371/journal.pone.0028228
Bioinformatic analysis and post-translational modification crosstalk prediction of lysine acetylation
Abstract
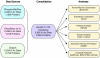
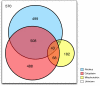
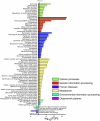
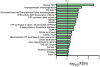
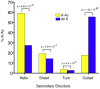
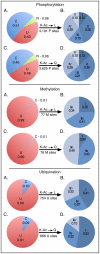
Recent proteomics studies suggest high abundance and a much wider role for lysine acetylation (K-Ac) in cellular functions. Nevertheless, cross influence between K-Ac and other post-translational modifications (PTMs) has not been carefully examined. Here, we used a variety of bioinformatics tools to analyze several available K-Ac datasets. Using gene ontology databases, we demonstrate that K-Ac sites are found in all cellular compartments. KEGG analysis indicates that the K-Ac sites are found on proteins responsible for a diverse and wide array of vital cellular functions. Domain structure prediction shows that K-Ac sites are found throughout a wide variety of protein domains, including those in heat shock proteins and those involved in cell cycle functions and DNA repair. Secondary structure prediction proves that K-Ac sites are preferentially found in ordered structures such as alpha helices and beta sheets. Finally, by mutating K-Ac sites in silico and predicting the effect on nearby phosphorylation sites, we demonstrate that the majority of lysine acetylation sites have the potential to impact protein phosphorylation, methylation, and ubiquitination status. Our work validates earlier smaller-scale studies on the acetylome and demonstrates the importance of PTM crosstalk for regulation of cellular function.
Conflict of interest statement
Figures
Similar articles
-
Comprehensive proteome analyses of lysine acetylation in tea leaves by sensing nitrogen nutrition.BMC Genomics. 2018 Nov 26;19(1):840. doi: 10.1186/s12864-018-5250-4. BMC Genomics. 2018. PMID: 30477445 Free PMC article.
-
Identification, Quantification, and Site Localization of Protein Posttranslational Modifications via Mass Spectrometry-Based Proteomics.Adv Exp Med Biol. 2016;919:345-382. doi: 10.1007/978-3-319-41448-5_17. Adv Exp Med Biol. 2016. PMID: 27975226 Review.
-
Proteome-wide analysis of lysine acetylation in the plant pathogen Botrytis cinerea.Sci Rep. 2016 Jul 6;6:29313. doi: 10.1038/srep29313. Sci Rep. 2016. PMID: 27381557 Free PMC article.
-
Advances in Prediction of Posttranslational Modification Sites Known to Localize in Protein Supersecondary Structures.Methods Mol Biol. 2025;2870:117-151. doi: 10.1007/978-1-0716-4213-9_8. Methods Mol Biol. 2025. PMID: 39543034
-
Prediction of lysine post-translational modifications using bioinformatic tools.Essays Biochem. 2012;52:165-77. doi: 10.1042/bse0520165. Essays Biochem. 2012. PMID: 22708570 Review.
Cited by
-
PTMcode: a database of known and predicted functional associations between post-translational modifications in proteins.Nucleic Acids Res. 2013 Jan;41(Database issue):D306-11. doi: 10.1093/nar/gks1230. Epub 2012 Nov 28. Nucleic Acids Res. 2013. PMID: 23193284 Free PMC article.
-
Quantitative acetylome analysis reveals histone modifications that may predict prognosis in hepatitis B-related hepatocellular carcinoma.Clin Transl Med. 2021 Mar;11(3):e313. doi: 10.1002/ctm2.313. Clin Transl Med. 2021. PMID: 33783990 Free PMC article.
-
Plant mitochondrial retrograde signaling: post-translational modifications enter the stage.Front Plant Sci. 2012 Nov 12;3:253. doi: 10.3389/fpls.2012.00253. eCollection 2012. Front Plant Sci. 2012. PMID: 23162565 Free PMC article.
-
In silico analysis of protein Lys-N(𝜀)-acetylation in plants.Front Plant Sci. 2014 Aug 4;5:381. doi: 10.3389/fpls.2014.00381. eCollection 2014. Front Plant Sci. 2014. PMID: 25136347 Free PMC article. Review.
-
PTM-ssMP: A Web Server for Predicting Different Types of Post-translational Modification Sites Using Novel Site-specific Modification Profile.Int J Biol Sci. 2018 May 22;14(8):946-956. doi: 10.7150/ijbs.24121. eCollection 2018. Int J Biol Sci. 2018. PMID: 29989096 Free PMC article.
References
-
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. - PubMed
-
- Allfrey VG, Mirsky AE. Structural Modifications of Histones and their Possible Role in the Regulation of RNA Synthesis. Science. 1964;144:559. - PubMed
-
- Pazin MJ, Kadonaga JT. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. - PubMed
-
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. - PubMed
-
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

