The role of Dicer protein partners in the processing of microRNA precursors
- PMID: 22163034
- PMCID: PMC3232248
- DOI: 10.1371/journal.pone.0028548
The role of Dicer protein partners in the processing of microRNA precursors
Abstract
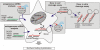
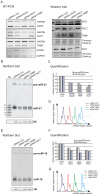
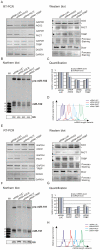
One of the cellular functions of the ribonuclease Dicer is to process microRNA precursors (pre-miRNAs) into mature microRNAs (miRNAs). Human Dicer performs this function in cooperation with its protein partners, AGO2, PACT and TRBP. The exact role of these accessory proteins in Dicer activity is still poorly understood. In this study, we used the northern blotting technique to investigate pre-miRNA cleavage efficiency and specificity after depletion of AGO2, PACT and TRBP by RNAi. The results showed that the inhibition of either Dicer protein partner substantially affected not only miRNA levels but also pre-miRNA levels, and it had a rather minor effect on the specificity of Dicer cleavage. The analysis of the Dicer cleavage products generated in vitro revealed the presence of a cleavage intermediate when pre-miRNA was processed by recombinant Dicer alone. This intermediate was not observed during pre-miRNA cleavage by endogenous Dicer. We demonstrate that AGO2, PACT and TRBP were required for the efficient functioning of Dicer in cells, and we suggest that one of the roles of these proteins is to assure better synchronization of cleavages triggered by two RNase III domains of Dicer.
Conflict of interest statement
Figures
Similar articles
-
Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing.Nucleic Acids Res. 2013 Jul;41(13):6568-76. doi: 10.1093/nar/gkt361. Epub 2013 May 9. Nucleic Acids Res. 2013. PMID: 23661684 Free PMC article.
-
Substrate-specific kinetics of Dicer-catalyzed RNA processing.J Mol Biol. 2010 Dec 3;404(3):392-402. doi: 10.1016/j.jmb.2010.09.030. Epub 2010 Oct 13. J Mol Biol. 2010. PMID: 20932845 Free PMC article.
-
Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis.Mol Cell. 2015 Feb 5;57(3):397-407. doi: 10.1016/j.molcel.2014.11.030. Epub 2014 Dec 31. Mol Cell. 2015. PMID: 25557550 Free PMC article.
-
Cryo-EM structures of human DICER dicing a pre-miRNA substrate.FEBS J. 2024 Jul;291(14):3072-3079. doi: 10.1111/febs.17048. Epub 2024 Jan 10. FEBS J. 2024. PMID: 38151772 Review.
-
The role of the precursor structure in the biogenesis of microRNA.Cell Mol Life Sci. 2011 Sep;68(17):2859-71. doi: 10.1007/s00018-011-0726-2. Epub 2011 May 24. Cell Mol Life Sci. 2011. PMID: 21607569 Free PMC article. Review.
Cited by
-
Regulation of miRNA biogenesis as an integrated component of growth factor signaling.Curr Opin Cell Biol. 2013 Apr;25(2):233-40. doi: 10.1016/j.ceb.2012.12.005. Epub 2013 Jan 8. Curr Opin Cell Biol. 2013. PMID: 23312066 Free PMC article. Review.
-
Sequence features of Drosha and Dicer cleavage sites affect the complexity of isomiRs.Int J Mol Sci. 2015 Apr 10;16(4):8110-27. doi: 10.3390/ijms16048110. Int J Mol Sci. 2015. PMID: 25867481 Free PMC article.
-
Adipose tissue macrophages secrete small extracellular vesicles that mediate rosiglitazone-induced insulin sensitization.Nat Metab. 2024 May;6(5):880-898. doi: 10.1038/s42255-024-01023-w. Epub 2024 Apr 11. Nat Metab. 2024. PMID: 38605183 Free PMC article.
-
Dicer knockdown inhibits endothelial cell tumor growth via microRNA 21a-3p targeting of Nox-4.J Biol Chem. 2014 Mar 28;289(13):9027-38. doi: 10.1074/jbc.M113.519264. Epub 2014 Feb 4. J Biol Chem. 2014. PMID: 24497637 Free PMC article.
-
Dicer in immune cell development and function.Immunol Invest. 2014;43(2):182-95. doi: 10.3109/08820139.2013.863557. Epub 2013 Dec 4. Immunol Invest. 2014. PMID: 24303839 Free PMC article. Review.
References
-
- MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr Opin Struct Biol. 2007;17:138–145. - PubMed
-
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

