PTEN modulates miR-21 processing via RNA-regulatory protein RNH1
- PMID: 22162762
- PMCID: PMC3230587
- DOI: 10.1371/journal.pone.0028308
PTEN modulates miR-21 processing via RNA-regulatory protein RNH1
Abstract
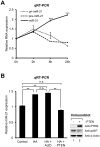
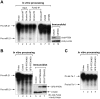
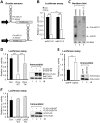
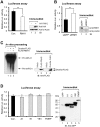
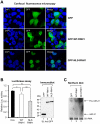
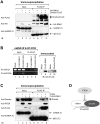
Aberrant miR-21 expression is closely associated with cell proliferation, anti-apoptosis, migration, invasion, and metastasis in various cancers. However, the regulatory mechanism of miR-21 biogenesis is largely unknown. Here, we demonstrated that the tumor suppressor PTEN negatively regulates the expression of oncogenic miR-21 at the post-transcriptional level. Moreover, our results suggest that PTEN plays such a role through the indirect interaction with the Drosha complex. To elucidate how PTEN regulates pri- to pre-miR-21 processing, we attempted to find PTEN-interacting proteins and identified an RNA-regulatory protein, RNH1. Using the sensor to monitor pri-miR-21 processing, we demonstrated that RNH1 is necessary and sufficient for pri-miR-21 processing. Moreover, our results propose that the nuclear localization of RNH1 is important for this function. Further analysis showed that RNH1 directly interacts with the Drosha complex and that PTEN blocks this interaction. Taken together, these results suggest that the PTEN-mediated miR-21 regulation is achieved by inhibiting the interaction between the Drosha complex and RNH1, revealing previously unidentified role of PTEN in the oncogenic miR-21 biogenesis.
Conflict of interest statement
Figures

Similar articles
-
SMAD proteins control DROSHA-mediated microRNA maturation.Nature. 2008 Jul 3;454(7200):56-61. doi: 10.1038/nature07086. Epub 2008 Jun 11. Nature. 2008. PMID: 18548003 Free PMC article.
-
MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer.Gastroenterology. 2007 Aug;133(2):647-58. doi: 10.1053/j.gastro.2007.05.022. Epub 2007 May 21. Gastroenterology. 2007. PMID: 17681183 Free PMC article.
-
Post-transcriptional modulation of protein phosphatase PPP2CA and tumor suppressor PTEN by endogenous siRNA cleaved from hairpin within PTEN mRNA 3'UTR in human liver cells.Acta Pharmacol Sin. 2016 Jul;37(7):898-907. doi: 10.1038/aps.2016.18. Epub 2016 May 2. Acta Pharmacol Sin. 2016. PMID: 27133296 Free PMC article.
-
MicroRNA-21 (Mir-21) Promotes Cell Growth and Invasion by Repressing Tumor Suppressor PTEN in Colorectal Cancer.Cell Physiol Biochem. 2017;43(3):945-958. doi: 10.1159/000481648. Epub 2017 Sep 29. Cell Physiol Biochem. 2017. PMID: 28957811
-
LncRNA TUSC8 inhibits the invasion and migration of cervical cancer cells via miR-641/PTEN axis.Cell Biol Int. 2019 Jul;43(7):781-788. doi: 10.1002/cbin.11152. Epub 2019 May 20. Cell Biol Int. 2019. PMID: 31033083
Cited by
-
Mechanical stretch modulates microRNA 21 expression, participating in proliferation and apoptosis in cultured human aortic smooth muscle cells.PLoS One. 2012;7(10):e47657. doi: 10.1371/journal.pone.0047657. Epub 2012 Oct 17. PLoS One. 2012. PMID: 23082189 Free PMC article.
-
Chromatolysis: Do injured axons regenerate poorly when ribonucleases attack rough endoplasmic reticulum, ribosomes and RNA?Dev Neurobiol. 2018 Oct;78(10):1011-1024. doi: 10.1002/dneu.22625. Epub 2018 Aug 1. Dev Neurobiol. 2018. PMID: 30027624 Free PMC article. Review.
-
Defining fallopian tube-derived miRNA cancer signatures.Cancer Med. 2019 Nov;8(15):6709-6716. doi: 10.1002/cam4.2416. Epub 2019 Sep 10. Cancer Med. 2019. PMID: 31503420 Free PMC article.
-
Negative regulation of lncRNA GAS5 by miR-21.Cell Death Differ. 2013 Nov;20(11):1558-68. doi: 10.1038/cdd.2013.110. Epub 2013 Aug 9. Cell Death Differ. 2013. PMID: 23933812 Free PMC article.
-
Ribonuclease Inhibitor 1 (RNH1) Regulates Sperm tsRNA Generation for Paternal Inheritance through Interacting with Angiogenin in the Caput Epididymis.Antioxidants (Basel). 2024 Aug 22;13(8):1020. doi: 10.3390/antiox13081020. Antioxidants (Basel). 2024. PMID: 39199264 Free PMC article.
References
-
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. - PubMed
-
- Garofalo M, Croce CM. microRNAs: Master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol. 2011;51:25–43. - PubMed
-
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

