Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity
- PMID: 22157767
- PMCID: PMC3285339
- DOI: 10.1074/jbc.M111.275057
Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity
Abstract
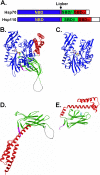
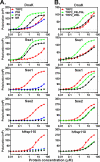
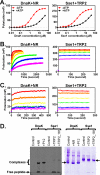
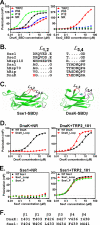
The molecular chaperone 70-kDa heat-shock proteins (Hsp70s) play essential roles in maintaining protein homeostasis. Hsp110, an Hsp70 homolog, is highly efficient in preventing protein aggregation but lacks the hallmark folding activity seen in Hsp70s. To understand the mechanistic differences between these two chaperones, we first characterized the distinct peptide substrate binding properties of Hsp110s. In contrast to Hsp70s, Hsp110s prefer aromatic residues in their substrates, and the substrate binding and release exhibit remarkably fast kinetics. Sequence and structure comparison revealed significant differences in the two peptide-binding loops: the length and properties are switched. When we swapped these two loops in an Hsp70, the peptide binding properties of this mutant Hsp70 were converted to Hsp110-like, and more impressively, it functionally behaved like an Hsp110. Thus, the peptide substrate binding properties implemented in the peptide-binding loops may determine the chaperone activity differences between Hsp70s and Hsp110s.
Figures

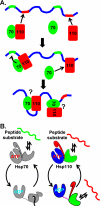
Similar articles
-
Interdomain interactions dictate the function of the Candida albicans Hsp110 protein Msi3.J Biol Chem. 2021 Sep;297(3):101082. doi: 10.1016/j.jbc.2021.101082. Epub 2021 Aug 14. J Biol Chem. 2021. PMID: 34403698 Free PMC article.
-
Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1.Cell. 2007 Oct 5;131(1):106-20. doi: 10.1016/j.cell.2007.08.039. Cell. 2007. PMID: 17923091 Free PMC article.
-
Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding.J Biol Chem. 2005 Dec 16;280(50):41252-61. doi: 10.1074/jbc.M503615200. Epub 2005 Oct 11. J Biol Chem. 2005. PMID: 16219770
-
The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s.Cell Stress Chaperones. 2000 Oct;5(4):276-90. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2. Cell Stress Chaperones. 2000. PMID: 11048651 Free PMC article. Review.
-
The Role of Non-Canonical Hsp70s (Hsp110/Grp170) in Cancer.Cells. 2021 Jan 28;10(2):254. doi: 10.3390/cells10020254. Cells. 2021. PMID: 33525518 Free PMC article. Review.
Cited by
-
Purification and biochemical characterization of Msi3, an essential Hsp110 molecular chaperone in Candida albicans.Cell Stress Chaperones. 2021 Jul;26(4):695-704. doi: 10.1007/s12192-021-01213-5. Epub 2021 May 28. Cell Stress Chaperones. 2021. PMID: 34047887 Free PMC article.
-
Alterations in Histone Methylation States Increased Profusion of Lethal(2)-Essential-for-Life-Like (l(2)elf), Trithorax and Polycomb Genes in Apis mellifera under Heat Stress.Insects. 2024 Jan 5;15(1):33. doi: 10.3390/insects15010033. Insects. 2024. PMID: 38249039 Free PMC article.
-
A novel and unique ATP hydrolysis to AMP by a human Hsp70 Binding immunoglobin protein (BiP).Protein Sci. 2022 Apr;31(4):797-810. doi: 10.1002/pro.4267. Epub 2021 Dec 31. Protein Sci. 2022. PMID: 34941000 Free PMC article.
-
Extrapolation of Inter Domain Communications and Substrate Binding Cavity of Camel HSP70 1A: A Molecular Modeling and Dynamics Simulation Study.PLoS One. 2015 Aug 27;10(8):e0136630. doi: 10.1371/journal.pone.0136630. eCollection 2015. PLoS One. 2015. PMID: 26313938 Free PMC article.
-
Mitochondrial HSP70 Chaperone System-The Influence of Post-Translational Modifications and Involvement in Human Diseases.Int J Mol Sci. 2021 Jul 28;22(15):8077. doi: 10.3390/ijms22158077. Int J Mol Sci. 2021. PMID: 34360841 Free PMC article. Review.
References
-
- Bukau B., Weissman J., Horwich A. (2006) Molecular chaperones and protein quality control. Cell 125, 443–451 - PubMed
-
- Hartl F. U., Hayer-Hartl M. (2009) Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16, 574–581 - PubMed
-
- Mayer M. P. (2010) Gymnastics of molecular chaperones. Mol. Cell 39, 321–331 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

