Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies
- PMID: 22152478
- PMCID: PMC3240842
- DOI: 10.1016/j.molcel.2011.09.019
Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies
Abstract
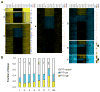
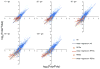
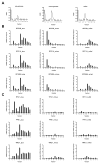
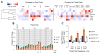
Cell survival in changing environments requires appropriate regulation of gene expression, including posttranscriptional regulatory mechanisms. From reporter gene studies in glucose-starved yeast, it was proposed that translationally silenced eukaryotic mRNAs accumulate in P bodies and can return to active translation. We present evidence contradicting the notion that reversible storage of nontranslating mRNAs is a widespread and general phenomenon. First, genome-wide measurements of mRNA abundance, translation, and ribosome occupancy after glucose withdrawal show that most mRNAs are depleted from the cell coincident with their depletion from polysomes. Second, only a limited subpopulation of translationally repressed transcripts, comprising fewer than 400 genes, can be reactivated for translation upon glucose readdition in the absence of new transcription. This highly selective posttranscriptional regulation could be a mechanism for cells to minimize the energetic costs of reversing gene-regulatory decisions in rapidly changing environments by transiently preserving a pool of transcripts whose translation is rate-limiting for growth.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies.Science. 2005 Oct 21;310(5747):486-9. doi: 10.1126/science.1115791. Epub 2005 Sep 1. Science. 2005. PMID: 16141371 Free PMC article.
-
Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae.Mol Biol Cell. 2007 Jul;18(7):2592-602. doi: 10.1091/mbc.e06-12-1149. Epub 2007 May 2. Mol Biol Cell. 2007. PMID: 17475768 Free PMC article.
-
The HOG pathway dictates the short-term translational response after hyperosmotic shock.Mol Biol Cell. 2010 Sep 1;21(17):3080-92. doi: 10.1091/mbc.E10-01-0006. Epub 2010 Jun 29. Mol Biol Cell. 2010. PMID: 20587780 Free PMC article.
-
Inhibition of translation initiation following glucose depletion in yeast facilitates a rationalization of mRNA content.Biochem Soc Trans. 2010 Aug;38(4):1131-6. doi: 10.1042/BST0381131. Biochem Soc Trans. 2010. PMID: 20659017 Review.
-
Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs.Curr Opin Cell Biol. 2009 Jun;21(3):403-8. doi: 10.1016/j.ceb.2009.03.005. Epub 2009 Apr 23. Curr Opin Cell Biol. 2009. PMID: 19394210 Free PMC article. Review.
Cited by
-
The role of stress-activated RNA-protein granules in surviving adversity.RNA. 2021 Apr 30;27(7):753-62. doi: 10.1261/rna.078738.121. Online ahead of print. RNA. 2021. PMID: 33931500 Free PMC article.
-
Granules harboring translationally active mRNAs provide a platform for P-body formation following stress.Cell Rep. 2014 Nov 6;9(3):944-54. doi: 10.1016/j.celrep.2014.09.040. Epub 2014 Oct 23. Cell Rep. 2014. PMID: 25437551 Free PMC article.
-
Genome-Wide Mapping of Decay Factor-mRNA Interactions in Yeast Identifies Nutrient-Responsive Transcripts as Targets of the Deadenylase Ccr4.G3 (Bethesda). 2018 Jan 4;8(1):315-330. doi: 10.1534/g3.117.300415. G3 (Bethesda). 2018. PMID: 29158339 Free PMC article.
-
Yeast Gis2 and its human ortholog CNBP are novel components of stress-induced RNP granules.PLoS One. 2012;7(12):e52824. doi: 10.1371/journal.pone.0052824. Epub 2012 Dec 21. PLoS One. 2012. PMID: 23285195 Free PMC article.
-
Droplet organelles?EMBO J. 2016 Aug 1;35(15):1603-12. doi: 10.15252/embj.201593517. Epub 2016 Jun 29. EMBO J. 2016. PMID: 27357569 Free PMC article. Review.
References
-
- Blake WJ, KAErn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

