Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3)
- PMID: 22135459
- PMCID: PMC3251073
- DOI: 10.1073/pnas.1116821108
Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3)
Abstract
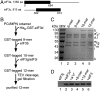
Protein fate in higher eukaryotes is controlled by three complexes that share conserved architectural elements: the proteasome, COP9 signalosome, and eukaryotic translation initiation factor 3 (eIF3). Here we reconstitute the 13-subunit human eIF3 in Escherichia coli, revealing its structural core to be the eight subunits with conserved orthologues in the proteasome lid complex and COP9 signalosome. This structural core in eIF3 binds to the small (40S) ribosomal subunit, to translation initiation factors involved in mRNA cap-dependent initiation, and to the hepatitis C viral (HCV) internal ribosome entry site (IRES) RNA. Addition of the remaining eIF3 subunits enables reconstituted eIF3 to assemble intact initiation complexes with the HCV IRES. Negative-stain EM reconstructions of reconstituted eIF3 further reveal how the approximately 400 kDa molecular mass structural core organizes the highly flexible 800 kDa molecular mass eIF3 complex, and mediates translation initiation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
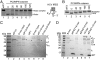



Similar articles
-
Structural roles for human translation factor eIF3 in initiation of protein synthesis.Science. 2005 Dec 2;310(5753):1513-5. doi: 10.1126/science.1118977. Science. 2005. PMID: 16322461
-
Unlike for cellular mRNAs and other viral internal ribosome entry sites (IRESs), the eIF3 subunit e is not required for the translational activity of the HCV IRES.J Biol Chem. 2020 Feb 14;295(7):1843-1856. doi: 10.1074/jbc.RA119.009502. Epub 2020 Jan 12. J Biol Chem. 2020. PMID: 31929110 Free PMC article.
-
Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit.Nature. 2013 Nov 28;503(7477):539-43. doi: 10.1038/nature12658. Epub 2013 Nov 3. Nature. 2013. PMID: 24185006 Free PMC article.
-
Hepatitis C Virus Translation Regulation.Int J Mol Sci. 2020 Mar 27;21(7):2328. doi: 10.3390/ijms21072328. Int J Mol Sci. 2020. PMID: 32230899 Free PMC article. Review.
-
Structural and mechanistic insights into hepatitis C viral translation initiation.Nat Rev Microbiol. 2007 Jan;5(1):29-38. doi: 10.1038/nrmicro1558. Epub 2006 Nov 27. Nat Rev Microbiol. 2007. PMID: 17128284 Review.
Cited by
-
The eIF3 complex of Leishmania-subunit composition and mode of recruitment to different cap-binding complexes.Nucleic Acids Res. 2015 Jul 27;43(13):6222-35. doi: 10.1093/nar/gkv564. Epub 2015 Jun 19. Nucleic Acids Res. 2015. PMID: 26092695 Free PMC article.
-
eIF3 interacts with histone H4 messenger RNA to regulate its translation.J Biol Chem. 2021 Jan-Jun;296:100578. doi: 10.1016/j.jbc.2021.100578. Epub 2021 Mar 23. J Biol Chem. 2021. PMID: 33766559 Free PMC article.
-
Phosphorylation stoichiometries of human eukaryotic initiation factors.Int J Mol Sci. 2014 Jun 27;15(7):11523-38. doi: 10.3390/ijms150711523. Int J Mol Sci. 2014. PMID: 24979134 Free PMC article.
-
Maternal separation with early weaning: a rodent model providing novel insights into neglect associated developmental deficits.Dev Psychopathol. 2012 Nov;24(4):1401-16. doi: 10.1017/S095457941200079X. Dev Psychopathol. 2012. PMID: 23062306 Free PMC article.
-
Functional and biochemical characterization of human eukaryotic translation initiation factor 3 in living cells.Mol Cell Biol. 2014 Aug;34(16):3041-52. doi: 10.1128/MCB.00663-14. Epub 2014 Jun 9. Mol Cell Biol. 2014. PMID: 24912683 Free PMC article.
References
-
- Pick E, Hofmann K, Glickman MH. PCI complexes: Beyond the proteasome, CSN, and eIF3 Troika. Mol Cell. 2009;35:260–264. - PubMed
-
- Sharon M, et al. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009;17:31–40. - PubMed
-
- Enchev RI, Schreiber A, Beuron F, Morris EP. Structural insights into the COP9 signalosome and its common architecture with the 26S proteasome lid and eIF3. Structure. 2010;18:518–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

