Novel transcript truncating function of Rap1p revealed by synthetic codon-optimized Ty1 retrotransposon
- PMID: 22135353
- PMCID: PMC3276619
- DOI: 10.1534/genetics.111.136648
Novel transcript truncating function of Rap1p revealed by synthetic codon-optimized Ty1 retrotransposon
Abstract
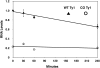
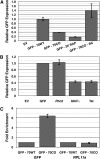
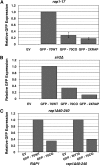
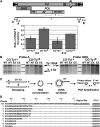
Extensive mutagenesis via massive recoding of retrotransposon Ty1 produced a synthetic codon-optimized retrotransposon (CO-Ty1). CO-Ty1 is defective for retrotransposition, suggesting a sequence capable of down-regulating retrotransposition. We mapped this sequence to a critical ~20-bp region within CO-Ty1 reverse transcriptase (RT) and confirmed that it reduced Ty1 transposition, protein, and RNA levels. Repression was not Ty1 specific; when introduced immediately downstream of the green fluorescent protein (GFP) stop codon, GFP expression was similarly reduced. Rap1p mediated this down-regulation, as shown by mutagenesis and chromatin immunoprecipitation. A regular threefold drop is observed in different contexts, suggesting utility for synthetic circuits. A large reduction of RNAP II occupancy on the CO-Ty1 construct was observed 3' to the identified Rap1p site and a novel 3' truncated RNA species was observed. We propose a novel mechanism of transcriptional regulation by Rap1p whereby it serves as a transcriptional roadblock when bound to transcription unit sequences.
Figures



Similar articles
-
The different (sur)faces of Rap1p.Mol Genet Genomics. 2003 Mar;268(6):791-8. doi: 10.1007/s00438-002-0801-3. Epub 2003 Jan 25. Mol Genet Genomics. 2003. PMID: 12655405 Review.
-
Remodeling yeast gene transcription by activating the Ty1 long terminal repeat retrotransposon under severe adenine deficiency.Mol Cell Biol. 2008 Sep;28(17):5543-54. doi: 10.1128/MCB.00416-08. Epub 2008 Jun 30. Mol Cell Biol. 2008. PMID: 18591253 Free PMC article.
-
A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon.Genome Res. 2012 Apr;22(4):704-13. doi: 10.1101/gr.129585.111. Epub 2012 Jan 4. Genome Res. 2012. PMID: 22219511 Free PMC article.
-
Differential effects of chromatin and Gcn4 on the 50-fold range of expression among individual yeast Ty1 retrotransposons.Mol Cell Biol. 2002 Apr;22(7):2078-88. doi: 10.1128/MCB.22.7.2078-2088.2002. Mol Cell Biol. 2002. PMID: 11884596 Free PMC article.
-
Reverse transcriptase and integrase of the Saccharomyces cerevisiae Ty1 element.Cytogenet Genome Res. 2005;110(1-4):269-87. doi: 10.1159/000084960. Cytogenet Genome Res. 2005. PMID: 16093680 Review.
Cited by
-
Repression of Divergent Noncoding Transcription by a Sequence-Specific Transcription Factor.Mol Cell. 2018 Dec 20;72(6):942-954.e7. doi: 10.1016/j.molcel.2018.10.018. Mol Cell. 2018. PMID: 30576656 Free PMC article.
-
Transcribe this way: Rap1 confers promoter directionality by repressing divergent transcription.Transcription. 2019 Jun;10(3):164-170. doi: 10.1080/21541264.2019.1608716. Epub 2019 May 5. Transcription. 2019. PMID: 31057041 Free PMC article. Review.
-
Artificial repressors for controlling gene expression in bacteria.Chem Commun (Camb). 2013 May 14;49(39):4325-7. doi: 10.1039/c2cc37107c. Epub 2012 Dec 10. Chem Commun (Camb). 2013. PMID: 23230569 Free PMC article.
-
Bug mapping and fitness testing of chemically synthesized chromosome X.Science. 2017 Mar 10;355(6329):eaaf4706. doi: 10.1126/science.aaf4706. Science. 2017. PMID: 28280152 Free PMC article.
-
Retrotransposon Ty1 RNA contains a 5'-terminal long-range pseudoknot required for efficient reverse transcription.RNA. 2013 Mar;19(3):320-32. doi: 10.1261/rna.035535.112. Epub 2013 Jan 17. RNA. 2013. PMID: 23329695 Free PMC article.
References
-
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R., 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154: 164–175 - PubMed
-
- Braiterman L. T., Monokian G. M., Eichinger D. J., Merbs S. L., Gabriel A., et al. , 1994. In-frame linker insertion mutagenesis of yeast transposon Ty1: phenotypic analysis. Gene 139: 19–26 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

