TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support
- PMID: 22135297
- PMCID: PMC3245116
- DOI: 10.1093/nar/gkr1161
TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support
Abstract
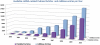
As the relevant literature and the number of experiments increase at a super linear rate, databases that curate and collect experimentally verified microRNA (miRNA) targets have gradually emerged. These databases attempt to provide efficient access to this wealth of experimental data, which is scattered in thousands of manuscripts. Aim of TarBase 6.0 (http://www.microrna.gr/tarbase) is to face this challenge by providing a significant increase of available miRNA targets derived from all contemporary experimental techniques (gene specific and high-throughput), while incorporating a powerful set of tools in a user-friendly interface. TarBase 6.0 hosts detailed information for each miRNA-gene interaction, ranging from miRNA- and gene-related facts to information specific to their interaction, the experimental validation methodologies and their outcomes. All database entries are enriched with function-related data, as well as general information derived from external databases such as UniProt, Ensembl and RefSeq. DIANA microT miRNA target prediction scores and the relevant prediction details are available for each interaction. TarBase 6.0 hosts the largest collection of manually curated experimentally validated miRNA-gene interactions (more than 65,000 targets), presenting a 16.5-175-fold increase over other available manually curated databases.
Figures

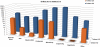

Similar articles
-
DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions.Nucleic Acids Res. 2015 Jan;43(Database issue):D153-9. doi: 10.1093/nar/gku1215. Epub 2014 Nov 21. Nucleic Acids Res. 2015. PMID: 25416803 Free PMC article.
-
DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions.Nucleic Acids Res. 2018 Jan 4;46(D1):D239-D245. doi: 10.1093/nar/gkx1141. Nucleic Acids Res. 2018. PMID: 29156006 Free PMC article.
-
DIANA-TarBase and DIANA Suite Tools: Studying Experimentally Supported microRNA Targets.Curr Protoc Bioinformatics. 2016 Sep 7;55:12.14.1-12.14.18. doi: 10.1002/cpbi.12. Curr Protoc Bioinformatics. 2016. PMID: 27603020
-
Review of databases for experimentally validated human microRNA-mRNA interactions.Database (Oxford). 2023 Apr 25;2023:baad014. doi: 10.1093/database/baad014. Database (Oxford). 2023. PMID: 37098414 Free PMC article. Review.
-
Web resources for microRNA research.Adv Exp Med Biol. 2013;774:225-50. doi: 10.1007/978-94-007-5590-1_12. Adv Exp Med Biol. 2013. PMID: 23377976 Review.
Cited by
-
An arrayed genome-scale lentiviral-enabled short hairpin RNA screen identifies lethal and rescuer gene candidates.Assay Drug Dev Technol. 2013 Apr;11(3):173-90. doi: 10.1089/adt.2012.475. Epub 2012 Nov 30. Assay Drug Dev Technol. 2013. PMID: 23198867 Free PMC article.
-
Dissecting the roles of microRNAs in coronary heart disease via integrative genomic analyses.Arterioscler Thromb Vasc Biol. 2015 Apr;35(4):1011-21. doi: 10.1161/ATVBAHA.114.305176. Epub 2015 Feb 5. Arterioscler Thromb Vasc Biol. 2015. PMID: 25657313 Free PMC article.
-
Principles of miRNA-mRNA interactions: beyond sequence complementarity.Cell Mol Life Sci. 2015 Aug;72(16):3127-41. doi: 10.1007/s00018-015-1922-2. Epub 2015 Jun 3. Cell Mol Life Sci. 2015. PMID: 26037721 Free PMC article. Review.
-
MREdictor: a two-step dynamic interaction model that accounts for mRNA accessibility and Pumilio binding accurately predicts microRNA targets.Nucleic Acids Res. 2013 Oct;41(18):8421-33. doi: 10.1093/nar/gkt629. Epub 2013 Jul 17. Nucleic Acids Res. 2013. PMID: 23863844 Free PMC article.
-
miRGate: a curated database of human, mouse and rat miRNA-mRNA targets.Database (Oxford). 2015 Apr 8;2015:bav035. doi: 10.1093/database/bav035. Print 2015. Database (Oxford). 2015. PMID: 25858286 Free PMC article.
References
-
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. - PubMed
-
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. - PubMed
-
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

