Plastid proteome assembly without Toc159: photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins
- PMID: 22128122
- PMCID: PMC3246318
- DOI: 10.1105/tpc.111.092882
Plastid proteome assembly without Toc159: photosynthetic protein import and accumulation of N-acetylated plastid precursor proteins
Abstract
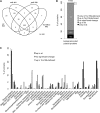
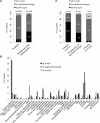
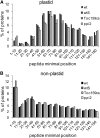
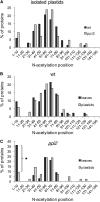
Import of nuclear-encoded precursor proteins from the cytosol is an essential step in chloroplast biogenesis that is mediated by protein translocon complexes at the inner and outer envelope membrane (TOC). Toc159 is thought to be the main receptor for photosynthetic proteins, but lacking a large-scale systems approach, this hypothesis has only been tested for a handful of photosynthetic and nonphotosynthetic proteins. To assess Toc159 precursor specificity, we quantitatively analyzed the accumulation of plastid proteins in two mutant lines deficient in this receptor. Parallel genome-wide transcript profiling allowed us to discern the consequences of impaired protein import from systemic transcriptional responses that contribute to the loss of photosynthetic capacity. On this basis, we defined putative Toc159-independent and Toc159-dependent precursor proteins. Many photosynthetic proteins accumulate in Toc159-deficient plastids, and, surprisingly, several distinct metabolic pathways are negatively affected by Toc159 depletion. Lack of Toc159 furthermore affects several proteins that accumulate as unprocessed N-acetylated precursor proteins outside of plastids. Together, our data show an unexpected client protein promiscuity of Toc159 that requires a far more differentiated view of Toc159 receptor function and regulation of plastid protein import, in which cytosolic Met removal followed by N-terminal acetylation of precursors emerges as an additional regulatory step.
Figures




Similar articles
-
Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors.Plant Cell. 2004 Aug;16(8):2059-77. doi: 10.1105/tpc.104.023309. Epub 2004 Jul 23. Plant Cell. 2004. PMID: 15273297 Free PMC article.
-
The chloroplast import receptor Toc90 partially restores the accumulation of Toc159 client proteins in the Arabidopsis thaliana ppi2 mutant.Mol Plant. 2011 Mar;4(2):252-63. doi: 10.1093/mp/ssq071. Epub 2011 Jan 10. Mol Plant. 2011. PMID: 21220583
-
A mutant of the Arabidopsis thaliana TOC159 gene accumulates reduced levels of linolenic acid and monogalactosyldiacylglycerol.Plant Physiol Biochem. 2013 Dec;73:344-50. doi: 10.1016/j.plaphy.2013.10.018. Epub 2013 Oct 21. Plant Physiol Biochem. 2013. PMID: 24184455
-
Protein transport in organelles: The Toc complex way of preprotein import.FEBS J. 2009 Mar;276(5):1156-65. doi: 10.1111/j.1742-4658.2009.06873.x. FEBS J. 2009. PMID: 19187236 Review.
-
Differentiation of chromoplasts and other plastids in plants.Plant Cell Rep. 2019 Jul;38(7):803-818. doi: 10.1007/s00299-019-02420-2. Epub 2019 May 11. Plant Cell Rep. 2019. PMID: 31079194 Free PMC article. Review.
Cited by
-
Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis.Mol Cell Proteomics. 2012 Sep;11(9):724-44. doi: 10.1074/mcp.M112.019208. Epub 2012 Jun 8. Mol Cell Proteomics. 2012. PMID: 22683509 Free PMC article.
-
Extrachloroplastic PP7L Functions in Chloroplast Development and Abiotic Stress Tolerance.Plant Physiol. 2019 May;180(1):323-341. doi: 10.1104/pp.19.00070. Epub 2019 Feb 13. Plant Physiol. 2019. PMID: 30760637 Free PMC article.
-
The Role of Tetrapyrrole- and GUN1-Dependent Signaling on Chloroplast Biogenesis.Plants (Basel). 2021 Jan 21;10(2):196. doi: 10.3390/plants10020196. Plants (Basel). 2021. PMID: 33494334 Free PMC article. Review.
-
Chloroplast Engineering: Fundamental Insights and Its Application in Amelioration of Environmental Stress.Appl Biochem Biotechnol. 2023 Apr;195(4):2463-2482. doi: 10.1007/s12010-022-03930-8. Epub 2022 Apr 28. Appl Biochem Biotechnol. 2023. PMID: 35484466 Review.
-
Importance of Translocon Subunit Tic56 for rRNA Processing and Chloroplast Ribosome Assembly.Plant Physiol. 2016 Dec;172(4):2429-2444. doi: 10.1104/pp.16.01393. Epub 2016 Oct 12. Plant Physiol. 2016. PMID: 27733515 Free PMC article.
References
-
- Agne B., Kessler F. (2009). Protein transport in organelles: The Toc complex way of preprotein import. FEBS J. 276: 1156–1165 - PubMed
-
- Baerenfaller K., Grossmann J., Grobei M.A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008). Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 - PubMed
-
- Baginsky S., Gruissem W. (2009). The chloroplast kinase network: new insights from large-scale phosphoproteome profiling. Mol. Plant 2: 1141–1153 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

