miR-150 regulates the development of NK and iNKT cells
- PMID: 22124110
- PMCID: PMC3244033
- DOI: 10.1084/jem.20111386
miR-150 regulates the development of NK and iNKT cells
Abstract
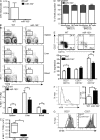
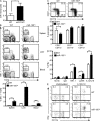
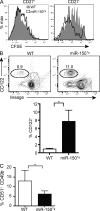
Natural killer (NK) and invariant NK T (iNKT) cells are critical in host defense against pathogens and for the initiation of adaptive immune responses. miRNAs play important roles in NK and iNKT cell development, maturation, and function, but the roles of specific miRNAs are unclear. We show that modulation of miR-150 expression levels has a differential effect on NK and iNKT cell development. Mice with a targeted deletion of miR-150 have an impaired, cell lineage-intrinsic defect in their ability to generate mature NK cells. Conversely, a gain-of-function miR-150 transgene promotes the development of NK cells, which display a more mature phenotype and are more responsive to activation. In contrast, overexpression of miR-150 results in a substantial reduction of iNKT cells in the thymus and in the peripheral lymphoid organs. The transcription factor c-Myb has been shown to be a direct target of miR-150. Our finding of increased NK cell and decreased iNKT cell frequencies in Myb heterozygous bone marrow chimeras suggests that miR-150 differentially controls the development of NK and iNKT cell lineages by targeting c-Myb.
Figures
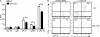



Similar articles
-
CD19-chimeric antigen receptor-invariant natural killer T cells transactivate NK cells and reduce alloreactivity.Cytotherapy. 2025 Jan;27(1):7-15. doi: 10.1016/j.jcyt.2024.08.004. Epub 2024 Aug 10. Cytotherapy. 2025. PMID: 39269404
-
B7H6 is the predominant activating ligand driving natural killer cell-mediated killing in patients with liquid tumours: evidence from clinical, in silico, in vitro, and in vivo studies.EBioMedicine. 2024 Dec;110:105459. doi: 10.1016/j.ebiom.2024.105459. Epub 2024 Nov 22. EBioMedicine. 2024. PMID: 39579618 Free PMC article.
-
The CML experience to elucidate the role of innate T-cells as effectors in the control of residual cancer cells and as potential targets for cancer therapy.Front Immunol. 2024 Nov 15;15:1473139. doi: 10.3389/fimmu.2024.1473139. eCollection 2024. Front Immunol. 2024. PMID: 39620210 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
MicroRNA Functions in Thymic Biology: Thymic Development and Involution.Front Immunol. 2018 Sep 11;9:2063. doi: 10.3389/fimmu.2018.02063. eCollection 2018. Front Immunol. 2018. PMID: 30254640 Free PMC article. Review.
-
Expression characteristics and interaction networks of microRNAs in spleen tissues of grass carp (Ctenopharyngodon idella).PLoS One. 2022 Mar 28;17(3):e0266189. doi: 10.1371/journal.pone.0266189. eCollection 2022. PLoS One. 2022. PMID: 35344574 Free PMC article.
-
Osteoporotic bone of miR-150-deficient mice: Possibly due to low serum OPG-mediated osteoclast activation.Bone Rep. 2015 Jun 23;3:5-10. doi: 10.1016/j.bonr.2015.06.003. eCollection 2015 Dec. Bone Rep. 2015. PMID: 28377961 Free PMC article.
-
Deregulated KLF4 Expression in Myeloid Leukemias Alters Cell Proliferation and Differentiation through MicroRNA and Gene Targets.Mol Cell Biol. 2015 Dec 7;36(4):559-73. doi: 10.1128/MCB.00712-15. Print 2016 Feb 15. Mol Cell Biol. 2015. PMID: 26644403 Free PMC article.
-
Regulated Expression of miR-155 is Required for iNKT Cell Development.Front Immunol. 2015 Mar 30;6:140. doi: 10.3389/fimmu.2015.00140. eCollection 2015. Front Immunol. 2015. PMID: 25870598 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

