Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms
- PMID: 22118463
- PMCID: PMC3227872
- DOI: 10.1016/j.cell.2011.10.039
Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms
Abstract
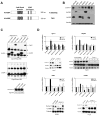
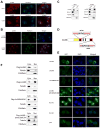
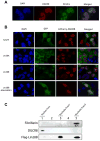
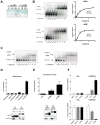
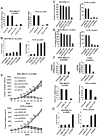
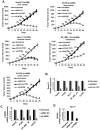
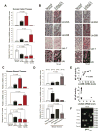
Lin28A and Lin28B selectively block the expression of let-7 microRNAs and function as oncogenes in a variety of human cancers. Lin28A recruits a TUTase (Zcchc11/TUT4) to let-7 precursors to block processing by Dicer in the cell cytoplasm. Here we find that unlike Lin28A, Lin28B represses let-7 processing through a Zcchc11-independent mechanism. Lin28B functions in the nucleus by sequestering primary let-7 transcripts and inhibiting their processing by the Microprocessor. The inhibitory effects of Zcchc11 depletion on the tumorigenic capacity and metastatic potential of human cancer cells and xenografts are restricted to Lin28A-expressing tumors. Furthermore, the majority of human colon and breast tumors analyzed exclusively express either Lin28A or Lin28B. Lin28A is expressed in HER2-overexpressing breast tumors, whereas Lin28B expression characterizes triple-negative breast tumors. Overall our results illuminate the distinct mechanisms by which Lin28A and Lin28B function and have implications for the development of new strategies for cancer therapy.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
A role of uridylation pathway for blockade of let-7 microRNA biogenesis by Lin28B.Cancer Sci. 2015 Sep;106(9):1174-81. doi: 10.1111/cas.12721. Epub 2015 Jul 14. Cancer Sci. 2015. PMID: 26080928 Free PMC article.
-
Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer.Mol Cancer. 2015 Jun 30;14:125. doi: 10.1186/s12943-015-0402-5. Mol Cancer. 2015. PMID: 26123544 Free PMC article. Review.
-
SET7/9 methylation of the pluripotency factor LIN28A is a nucleolar localization mechanism that blocks let-7 biogenesis in human ESCs.Cell Stem Cell. 2014 Dec 4;15(6):735-49. doi: 10.1016/j.stem.2014.10.016. Cell Stem Cell. 2014. PMID: 25479749 Free PMC article.
-
Comparison of the expression and function of Lin28A and Lin28B in colon cancer.Oncotarget. 2016 Nov 29;7(48):79605-79616. doi: 10.18632/oncotarget.12869. Oncotarget. 2016. PMID: 27793004 Free PMC article.
-
Progress on the role of LIN28A/B in tumor development and progression.Yi Chuan. 2024 Jun 20;46(6):452-465. doi: 10.16288/j.yczz.24-056. Yi Chuan. 2024. PMID: 38886149 Review.
Cited by
-
The tRNA pseudouridine synthase TruB1 regulates the maturation of let-7 miRNA.EMBO J. 2020 Oct 15;39(20):e104708. doi: 10.15252/embj.2020104708. Epub 2020 Sep 14. EMBO J. 2020. PMID: 32926445 Free PMC article.
-
MicroRNAs shape the neuronal landscape.Neuron. 2012 Aug 9;75(3):363-79. doi: 10.1016/j.neuron.2012.07.005. Neuron. 2012. PMID: 22884321 Free PMC article. Review.
-
Hepatitis B virus and microRNAs: Complex interactions affecting hepatitis B virus replication and hepatitis B virus-associated diseases.World J Gastroenterol. 2015 Jun 28;21(24):7375-99. doi: 10.3748/wjg.v21.i24.7375. World J Gastroenterol. 2015. PMID: 26139985 Free PMC article. Review.
-
LIN28 cooperates with WNT signaling to drive invasive intestinal and colorectal adenocarcinoma in mice and humans.Genes Dev. 2015 May 15;29(10):1074-86. doi: 10.1101/gad.256693.114. Epub 2015 May 8. Genes Dev. 2015. PMID: 25956904 Free PMC article.
-
Outside the coding genome, mammalian microRNAs confer structural and functional complexity.Sci Signal. 2015 Mar 17;8(368):re2. doi: 10.1126/scisignal.2005813. Sci Signal. 2015. PMID: 25783159 Free PMC article. Review.
References
-
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. - PubMed
-
- Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–36. - PubMed
-
- Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

