Phytozome: a comparative platform for green plant genomics
- PMID: 22110026
- PMCID: PMC3245001
- DOI: 10.1093/nar/gkr944
Phytozome: a comparative platform for green plant genomics
Abstract
The number of sequenced plant genomes and associated genomic resources is growing rapidly with the advent of both an increased focus on plant genomics from funding agencies, and the application of inexpensive next generation sequencing. To interact with this increasing body of data, we have developed Phytozome (http://www.phytozome.net), a comparative hub for plant genome and gene family data and analysis. Phytozome provides a view of the evolutionary history of every plant gene at the level of sequence, gene structure, gene family and genome organization, while at the same time providing access to the sequences and functional annotations of a growing number (currently 25) of complete plant genomes, including all the land plants and selected algae sequenced at the Joint Genome Institute, as well as selected species sequenced elsewhere. Through a comprehensive plant genome database and web portal, these data and analyses are available to the broader plant science research community, providing powerful comparative genomics tools that help to link model systems with other plants of economic and ecological importance.
Figures
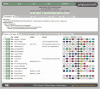
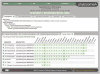
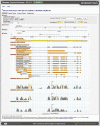
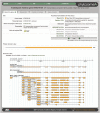
Similar articles
-
Closing target trimming and CTTdocker programs for discovering hidden superfamily loci in genomes.PLoS One. 2019 Jul 2;14(7):e0209468. doi: 10.1371/journal.pone.0209468. eCollection 2019. PLoS One. 2019. PMID: 31265455 Free PMC article.
-
PLAZA: a comparative genomics resource to study gene and genome evolution in plants.Plant Cell. 2009 Dec;21(12):3718-31. doi: 10.1105/tpc.109.071506. Epub 2009 Dec 29. Plant Cell. 2009. PMID: 20040540 Free PMC article.
-
Gramene 2013: comparative plant genomics resources.Nucleic Acids Res. 2014 Jan;42(Database issue):D1193-9. doi: 10.1093/nar/gkt1110. Epub 2013 Nov 11. Nucleic Acids Res. 2014. PMID: 24217918 Free PMC article.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
-
The Chlamydomonas genome project: a decade on.Trends Plant Sci. 2014 Oct;19(10):672-80. doi: 10.1016/j.tplants.2014.05.008. Epub 2014 Jun 17. Trends Plant Sci. 2014. PMID: 24950814 Free PMC article. Review.
Cited by
-
Genome-wide analysis and expression profile of the bZIP gene family in Neopyropia yezoensis.Front Plant Sci. 2024 Oct 21;15:1461922. doi: 10.3389/fpls.2024.1461922. eCollection 2024. Front Plant Sci. 2024. PMID: 39498397 Free PMC article.
-
Challenges in Assembling the Dated Tree of Life.Genome Biol Evol. 2024 Oct 9;16(10):evae229. doi: 10.1093/gbe/evae229. Genome Biol Evol. 2024. PMID: 39475308 Free PMC article.
-
Arabidopsis thaliana resistance to fusarium oxysporum 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection.PLoS Genet. 2013 May;9(5):e1003525. doi: 10.1371/journal.pgen.1003525. Epub 2013 May 23. PLoS Genet. 2013. PMID: 23717215 Free PMC article.
-
Genome-wide landscape of alternative splicing events in Brachypodium distachyon.DNA Res. 2013 Apr;20(2):163-71. doi: 10.1093/dnares/dss041. Epub 2013 Jan 7. DNA Res. 2013. PMID: 23297300 Free PMC article.
-
Analysis of Cell Wall-Related Genes in Organs of Medicago sativa L. under Different Abiotic Stresses.Int J Mol Sci. 2015 Jul 16;16(7):16104-24. doi: 10.3390/ijms160716104. Int J Mol Sci. 2015. PMID: 26193255 Free PMC article.
References
-
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. The sol genomics network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 2011;39:D1149–D1155. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

