High-resolution human cytomegalovirus transcriptome
- PMID: 22109557
- PMCID: PMC3241806
- DOI: 10.1073/pnas.1115861108
High-resolution human cytomegalovirus transcriptome
Abstract
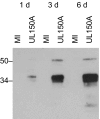
Deep sequencing was used to bring high resolution to the human cytomegalovirus (HCMV) transcriptome at the stage when infectious virion production is under way, and major findings were confirmed by extensive experimentation using conventional techniques. The majority (65.1%) of polyadenylated viral RNA transcription is committed to producing four noncoding transcripts (RNA2.7, RNA1.2, RNA4.9, and RNA5.0) that do not substantially overlap designated protein-coding regions. Additional noncoding RNAs that are transcribed antisense to protein-coding regions map throughout the genome and account for 8.7% of transcription from these regions. RNA splicing is more common than recognized previously, which was evidenced by the identification of 229 potential donor and 132 acceptor sites, and it affects 58 protein-coding genes. The great majority (94) of 96 splice junctions most abundantly represented in the deep-sequencing data was confirmed by RT-PCR or RACE or supported by involvement in alternative splicing. Alternative splicing is frequent and particularly evident in four genes (RL8A, UL74A, UL124, and UL150A) that are transcribed by splicing from any one of many upstream exons. The analysis also resulted in the annotation of four previously unrecognized protein-coding regions (RL8A, RL9A, UL150A, and US33A), and expression of the UL150A protein was shown in the context of HCMV infection. The overall conclusion, that HCMV transcription is complex and multifaceted, has implications for the potential sophistication of virus functionality during infection. The study also illustrates the key contribution that deep sequencing can make to the genomics of nuclear DNA viruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Characterisation of transcripts from the human cytomegalovirus genes TRL7, UL20a, UL36, UL65, UL94, US3 and US34.Virus Genes. 2002;24(1):39-48. doi: 10.1023/a:1014033920070. Virus Genes. 2002. PMID: 11928987
-
Human cytomegalovirus UL37 immediate early target minigene RNAs are accurately spliced and polyadenylated.J Gen Virol. 2003 Jan;84(Pt 1):29-39. doi: 10.1099/vir.0.18700-0. J Gen Virol. 2003. PMID: 12533698
-
Functional and molecular dissection of HCMV long non-coding RNAs.Sci Rep. 2022 Nov 11;12(1):19303. doi: 10.1038/s41598-022-23317-3. Sci Rep. 2022. PMID: 36369338 Free PMC article.
-
Insights into the Transcriptome of Human Cytomegalovirus: A Comprehensive Review.Viruses. 2023 Aug 8;15(8):1703. doi: 10.3390/v15081703. Viruses. 2023. PMID: 37632045 Free PMC article. Review.
-
Human CMV transcripts: an overview.Future Microbiol. 2012 May;7(5):577-93. doi: 10.2217/fmb.12.32. Future Microbiol. 2012. PMID: 22568714 Review.
Cited by
-
Analysis and mapping of a 3' coterminal transcription unit derived from human cytomegalovirus open reading frames UL30-UL32.Virol J. 2013 Feb 27;10:65. doi: 10.1186/1743-422X-10-65. Virol J. 2013. PMID: 23446136 Free PMC article.
-
Deep sequencing analysis of defective genomes of parainfluenza virus 5 and their role in interferon induction.J Virol. 2013 May;87(9):4798-807. doi: 10.1128/JVI.03383-12. Epub 2013 Feb 28. J Virol. 2013. PMID: 23449801 Free PMC article.
-
Comparative genomics of carp herpesviruses.J Virol. 2013 Mar;87(5):2908-22. doi: 10.1128/JVI.03206-12. Epub 2012 Dec 26. J Virol. 2013. PMID: 23269803 Free PMC article.
-
HCMV Infection and Apoptosis: How Do Monocytes Survive HCMV Infection?Viruses. 2018 Sep 29;10(10):533. doi: 10.3390/v10100533. Viruses. 2018. PMID: 30274264 Free PMC article. Review.
-
Engineering, decoding and systems-level characterization of chimpanzee cytomegalovirus.PLoS Pathog. 2022 Jan 4;18(1):e1010193. doi: 10.1371/journal.ppat.1010193. eCollection 2022 Jan. PLoS Pathog. 2022. PMID: 34982803 Free PMC article.
References
-
- Davison AJ. In: Human Herpesviruses: Biology, Therapy and Immunoprophylaxis. Arvin A, et al., editors. Cambridge, UK: Cambridge University Press; 2007. pp. 10–26. - PubMed
-
- Dolan A, et al. Genetic content of wild-type human cytomegalovirus. J Gen Virol. 2004;85:1301–1312. - PubMed
-
- Scalzo AA, Forbes CA, Smith LM, Loh LC. Transcriptional analysis of human cytomegalovirus and rat cytomegalovirus homologues of the M73/M73.5 spliced gene family. Arch Virol. 2009;154(1):65–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

