Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling
- PMID: 22102825
- PMCID: PMC3213160
- DOI: 10.1371/journal.pgen.1002363
Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling
Abstract
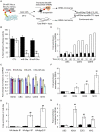
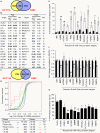
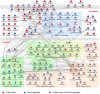
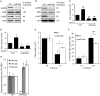
A simple biochemical method to isolate mRNAs pulled down with a transfected, biotinylated microRNA was used to identify direct target genes of miR-34a, a tumor suppressor gene. The method reidentified most of the known miR-34a regulated genes expressed in K562 and HCT116 cancer cell lines. Transcripts for 982 genes were enriched in the pull-down with miR-34a in both cell lines. Despite this large number, validation experiments suggested that ~90% of the genes identified in both cell lines can be directly regulated by miR-34a. Thus miR-34a is capable of regulating hundreds of genes. The transcripts pulled down with miR-34a were highly enriched for their roles in growth factor signaling and cell cycle progression. These genes form a dense network of interacting gene products that regulate multiple signal transduction pathways that orchestrate the proliferative response to external growth stimuli. Multiple candidate miR-34a-regulated genes participate in RAS-RAF-MAPK signaling. Ectopic miR-34a expression reduced basal ERK and AKT phosphorylation and enhanced sensitivity to serum growth factor withdrawal, while cells genetically deficient in miR-34a were less sensitive. Fourteen new direct targets of miR-34a were experimentally validated, including genes that participate in growth factor signaling (ARAF and PIK3R2) as well as genes that regulate cell cycle progression at various phases of the cell cycle (cyclins D3 and G2, MCM2 and MCM5, PLK1 and SMAD4). Thus miR-34a tempers the proliferative and pro-survival effect of growth factor stimulation by interfering with growth factor signal transduction and downstream pathways required for cell division.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
MicroRNA-34a is a tumor suppressor in choriocarcinoma via regulation of Delta-like1.BMC Cancer. 2013 Jan 18;13:25. doi: 10.1186/1471-2407-13-25. BMC Cancer. 2013. PMID: 23327670 Free PMC article.
-
MiR-34a targets GAS1 to promote cell proliferation and inhibit apoptosis in papillary thyroid carcinoma via PI3K/Akt/Bad pathway.Biochem Biophys Res Commun. 2013 Nov 29;441(4):958-63. doi: 10.1016/j.bbrc.2013.11.010. Epub 2013 Nov 9. Biochem Biophys Res Commun. 2013. PMID: 24220341
-
Hypoxia-induced down-regulation of microRNA-34a promotes EMT by targeting the Notch signaling pathway in tubular epithelial cells.PLoS One. 2012;7(2):e30771. doi: 10.1371/journal.pone.0030771. Epub 2012 Feb 17. PLoS One. 2012. PMID: 22363487 Free PMC article.
-
Expanding the Biotherapeutics Realm via miR-34a: "Potent Clever Little" Agent in Breast Cancer Therapy.Curr Pharm Biotechnol. 2019;20(8):665-673. doi: 10.2174/1389201020666190617162042. Curr Pharm Biotechnol. 2019. PMID: 31244419 Review.
-
The comprehensive landscape of miR-34a in cancer research.Cancer Metastasis Rev. 2021 Sep;40(3):925-948. doi: 10.1007/s10555-021-09973-3. Epub 2021 May 6. Cancer Metastasis Rev. 2021. PMID: 33959850 Review.
Cited by
-
LincRNA-p21 suppresses target mRNA translation.Mol Cell. 2012 Aug 24;47(4):648-55. doi: 10.1016/j.molcel.2012.06.027. Epub 2012 Jul 26. Mol Cell. 2012. PMID: 22841487 Free PMC article.
-
Embryonic stem cell miRNAs and their roles in development and disease.Semin Cancer Biol. 2012 Oct;22(5-6):428-36. doi: 10.1016/j.semcancer.2012.04.009. Epub 2012 May 4. Semin Cancer Biol. 2012. PMID: 22561239 Free PMC article. Review.
-
Deregulation of SATB2 in carcinogenesis with emphasis on miRNA-mediated control.Carcinogenesis. 2019 May 14;40(3):393-402. doi: 10.1093/carcin/bgz020. Carcinogenesis. 2019. PMID: 30916759 Free PMC article. Review.
-
An integrated miRNA functional screening and target validation method for organ morphogenesis.Sci Rep. 2016 Mar 16;6:23215. doi: 10.1038/srep23215. Sci Rep. 2016. PMID: 26980315 Free PMC article.
-
Plasma microRNA signature in presymptomatic and symptomatic subjects with C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis.J Neurol Neurosurg Psychiatry. 2021 May;92(5):485-493. doi: 10.1136/jnnp-2020-324647. Epub 2020 Nov 25. J Neurol Neurosurg Psychiatry. 2021. PMID: 33239440 Free PMC article.
References
-
- Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. - DOI - PubMed
-
- He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network [mdash] another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. - DOI - PMC - PubMed
-
- Sun F, Fu H, Liu Q, Tie Y, Zhu J, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Letters. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. - DOI - PubMed
-
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proceedings of the National Academy of Sciences. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. - DOI - PMC - PubMed
-
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proceedings of the National Academy of Sciences. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

