The human herpesvirus-7 (HHV-7) U21 immunoevasin subverts NK-mediated cytoxicity through modulation of MICA and MICB
- PMID: 22102813
- PMCID: PMC3213103
- DOI: 10.1371/journal.ppat.1002362
The human herpesvirus-7 (HHV-7) U21 immunoevasin subverts NK-mediated cytoxicity through modulation of MICA and MICB
Abstract
Herpesviruses have evolved numerous immune evasion strategies to facilitate establishment of lifelong persistent infections. Many herpesviruses encode gene products devoted to preventing viral antigen presentation as a means of escaping detection by cytotoxic T lymphocytes. The human herpesvirus-7 (HHV-7) U21 gene product, for example, is an immunoevasin that binds to class I major histocompatibility complex molecules and redirects them to the lysosomal compartment. Virus infection can also induce the upregulation of surface ligands that activate NK cells. Accordingly, the herpesviruses have evolved a diverse array of mechanisms to prevent NK cell engagement of NK-activating ligands on virus-infected cells. Here we demonstrate that the HHV-7 U21 gene product interferes with NK recognition. U21 can bind to the NK activating ligand ULBP1 and reroute it to the lysosomal compartment. In addition, U21 downregulates the surface expression of the NK activating ligands MICA and MICB, resulting in a reduction in NK-mediated cytotoxicity. These results suggest that this single viral protein may interfere both with CTL-mediated recognition through the downregulation of class I MHC molecules as well as NK-mediated recognition through downregulation of NK activating ligands.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



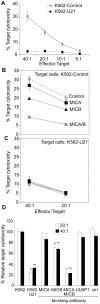
Similar articles
-
The HHV-6A Proteins U20 and U21 Target NKG2D Ligands to Escape Immune Recognition.Front Immunol. 2021 Oct 15;12:714799. doi: 10.3389/fimmu.2021.714799. eCollection 2021. Front Immunol. 2021. PMID: 34721381 Free PMC article.
-
Human herpesvirus 7 U21 tetramerizes to associate with class I major histocompatibility complex molecules.J Virol. 2014 Mar;88(6):3298-308. doi: 10.1128/JVI.02639-13. Epub 2014 Jan 3. J Virol. 2014. PMID: 24390327 Free PMC article.
-
Human herpesvirus 7 u21 downregulates classical and nonclassical class I major histocompatibility complex molecules from the cell surface.J Virol. 2010 Apr;84(8):3738-51. doi: 10.1128/JVI.01782-09. Epub 2010 Jan 27. J Virol. 2010. PMID: 20106916 Free PMC article.
-
Structural Models for Roseolovirus U20 And U21: Non-Classical MHC-I Like Proteins From HHV-6A, HHV-6B, and HHV-7.Front Immunol. 2022 Apr 4;13:864898. doi: 10.3389/fimmu.2022.864898. eCollection 2022. Front Immunol. 2022. PMID: 35444636 Free PMC article. Review.
-
The UL16-binding proteins, a novel family of MHC class I-related ligands for NKG2D, activate natural killer cell functions.Immunol Rev. 2001 Jun;181:185-92. doi: 10.1034/j.1600-065x.2001.1810115.x. Immunol Rev. 2001. PMID: 11513139 Review.
Cited by
-
Immune evasion by adenoviruses: a window into host-virus adaptation.FEBS Lett. 2019 Dec;593(24):3496-3503. doi: 10.1002/1873-3468.13682. Epub 2019 Dec 5. FEBS Lett. 2019. PMID: 31736048 Free PMC article. Review.
-
Human herpesviridae methods of natural killer cell evasion.Adv Virol. 2012;2012:359869. doi: 10.1155/2012/359869. Epub 2012 Jul 8. Adv Virol. 2012. PMID: 22829821 Free PMC article.
-
Characterization of the HHV-6B U20 Immunoevasin.J Virol. 2023 Feb 28;97(2):e0189022. doi: 10.1128/jvi.01890-22. Epub 2023 Jan 23. J Virol. 2023. PMID: 36688652 Free PMC article.
-
Convergent Evolution by Cancer and Viruses in Evading the NKG2D Immune Response.Cancers (Basel). 2020 Dec 18;12(12):3827. doi: 10.3390/cancers12123827. Cancers (Basel). 2020. PMID: 33352921 Free PMC article. Review.
-
Human NKG2D-ligands: cell biology strategies to ensure immune recognition.Front Immunol. 2012 Sep 25;3:299. doi: 10.3389/fimmu.2012.00299. eCollection 2012. Front Immunol. 2012. PMID: 23056001 Free PMC article.
References
-
- Tanaka-Taya K, Kondo T, Mukai T, Miyoshi H, Yamamoto Y, et al. Seroepidemiological study of human herpesvirus-6 and -7 in children of different ages and detection of these two viruses in throat swabs by polymerase chain reaction. J Med Virol. 1996;48:88–94. - PubMed
-
- Hansen TH, Bouvier M. MHC class I antigen presentation: learning from viral evasion strategies. Nat Rev Immunol. 2009;9:503–513. - PubMed
-
- Powers C, DeFilippis V, Malouli D, Fruh K. Cytomegalovirus immune evasion. Curr Top Microbiol Immunol. 2008;325:333–359. - PubMed
-
- Yewdell JW, Hill AB. Viral interference with antigen presentation. Nat Immunol. 2002;3:1019–1025. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

