Microplate-based platform for combined chromatin and DNA methylation immunoprecipitation assays
- PMID: 22098709
- PMCID: PMC3247195
- DOI: 10.1186/1471-2199-12-49
Microplate-based platform for combined chromatin and DNA methylation immunoprecipitation assays
Abstract
Background: The processes that compose expression of a given gene are far more complex than previously thought presenting unprecedented conceptual and mechanistic challenges that require development of new tools. Chromatin structure, which is regulated by DNA methylation and histone modification, is at the center of gene regulation. Immunoprecipitations of chromatin (ChIP) and methylated DNA (MeDIP) represent a major achievement in this area that allow researchers to probe chromatin modifications as well as specific protein-DNA interactions in vivo and to estimate the density of proteins at specific sites genome-wide. Although a critical component of chromatin structure, DNA methylation has often been studied independently of other chromatin events and transcription.
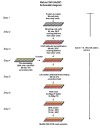
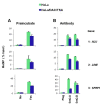
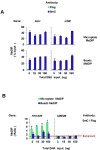
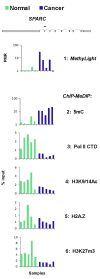
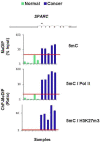
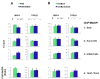
Results: To allow simultaneous measurements of DNA methylation with other genomic processes, we developed and validated a simple and easy-to-use high throughput microplate-based platform for analysis of DNA methylation. Compared to the traditional beads-based MeDIP the microplate MeDIP was more sensitive and had lower non-specific binding. We integrated the MeDIP method with a microplate ChIP assay which allows measurements of both DNA methylation and histone marks at the same time, Matrix ChIP-MeDIP platform. We illustrated several applications of this platform to relate DNA methylation, with chromatin and transcription events at selected genes in cultured cells, human cancer and in a model of diabetic kidney disease.
Conclusion: The high throughput capacity of Matrix ChIP-MeDIP to profile tens and potentially hundreds of different genomic events at the same time as DNA methylation represents a powerful platform to explore complex genomic mechanism at selected genes in cultured cells and in whole tissues. In this regard, Matrix ChIP-MeDIP should be useful to complement genome-wide studies where the rich chromatin and transcription database resources provide fruitful foundation to pursue mechanistic, functional and diagnostic information at genes of interest in health and disease.
Figures
Similar articles
-
Microplate-based chromatin immunoprecipitation method, Matrix ChIP: a platform to study signaling of complex genomic events.Nucleic Acids Res. 2008 Feb;36(3):e17. doi: 10.1093/nar/gkn001. Epub 2008 Jan 18. Nucleic Acids Res. 2008. PMID: 18203739 Free PMC article.
-
A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis.Nat Biotechnol. 2008 Jul;26(7):779-85. doi: 10.1038/nbt1414. Nat Biotechnol. 2008. PMID: 18612301 Free PMC article.
-
Genome-wide epigenetic analysis of human pluripotent stem cells by ChIP and ChIP-Seq.Methods Mol Biol. 2011;767:253-67. doi: 10.1007/978-1-61779-201-4_19. Methods Mol Biol. 2011. PMID: 21822881
-
Chop it, ChIP it, check it: the current status of chromatin immunoprecipitation.Front Biosci. 2008 Jan 1;13:929-43. doi: 10.2741/2733. Front Biosci. 2008. PMID: 17981601 Review.
-
Immunoprecipitation of methylated DNA.Methods Mol Biol. 2009;567:249-62. doi: 10.1007/978-1-60327-414-2_16. Methods Mol Biol. 2009. PMID: 19588097 Review.
Cited by
-
Distinct patterns of transcriptional and epigenetic alterations characterize acute and chronic kidney injury.Sci Rep. 2018 Dec 14;8(1):17870. doi: 10.1038/s41598-018-35943-x. Sci Rep. 2018. PMID: 30552397 Free PMC article.
-
Methylation-Demethylation Dynamics: Implications of Changes in Acute Kidney Injury.Anal Cell Pathol (Amst). 2018 Jul 8;2018:8764384. doi: 10.1155/2018/8764384. eCollection 2018. Anal Cell Pathol (Amst). 2018. PMID: 30073137 Free PMC article. Review.
-
Beads-free protein immunoprecipitation for a mass spectrometry-based interactome and posttranslational modifications analysis.Proteome Sci. 2015 Sep 2;13:23. doi: 10.1186/s12953-015-0079-0. eCollection 2015. Proteome Sci. 2015. PMID: 26336360 Free PMC article.
-
Regulation of ribosomal RNA expression across the lifespan is fine-tuned by maternal diet before implantation.Biochim Biophys Acta. 2016 Jul;1859(7):906-13. doi: 10.1016/j.bbagrm.2016.04.001. Epub 2016 Apr 7. Biochim Biophys Acta. 2016. PMID: 27060415 Free PMC article.
-
Chromatin changes trigger laminin genes dysregulation in aging kidneys.Aging (Albany NY). 2018 May 29;10(5):1133-1145. doi: 10.18632/aging.101453. Aging (Albany NY). 2018. PMID: 29846172 Free PMC article.
References
-
- Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

