Complex dynamic development of poliovirus membranous replication complexes
- PMID: 22072780
- PMCID: PMC3255921
- DOI: 10.1128/JVI.05937-11
Complex dynamic development of poliovirus membranous replication complexes
Abstract
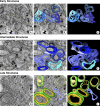
Replication of all positive-strand RNA viruses is intimately associated with membranes. Here we utilize electron tomography and other methods to investigate the remodeling of membranes in poliovirus-infected cells. We found that the viral replication structures previously described as "vesicles" are in fact convoluted, branching chambers with complex and dynamic morphology. They are likely to originate from cis-Golgi membranes and are represented during the early stages of infection by single-walled connecting and branching tubular compartments. These early viral organelles gradually transform into double-membrane structures by extension of membranous walls and/or collapsing of the luminal cavity of the single-membrane structures. As the double-membrane regions develop, they enclose cytoplasmic material. At this stage, a continuous membranous structure may have double- and single-walled membrane morphology at adjacent cross-sections. In the late stages of the replication cycle, the structures are represented mostly by double-membrane vesicles. Viral replication proteins, double-stranded RNA species, and actively replicating RNA are associated with both double- and single-membrane structures. However, the exponential phase of viral RNA synthesis occurs when single-membrane formations are predominant in the cell. It has been shown previously that replication complexes of some other positive-strand RNA viruses form on membrane invaginations, which result from negative membrane curvature. Our data show that the remodeling of cellular membranes in poliovirus-infected cells produces structures with positive curvature of membranes. Thus, it is likely that there is a fundamental divergence in the requirements for the supporting cellular membrane-shaping machinery among different groups of positive-strand RNA viruses.
Figures






Similar articles
-
Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis.J Virol. 2000 Jul;74(14):6570-80. doi: 10.1128/jvi.74.14.6570-6580.2000. J Virol. 2000. PMID: 10864671 Free PMC article.
-
Generation of unique poliovirus RNA replication organelles.mBio. 2014 Feb 25;5(2):e00833-13. doi: 10.1128/mBio.00833-13. mBio. 2014. PMID: 24570367 Free PMC article.
-
Purification of Highly Active Alphavirus Replication Complexes Demonstrates Altered Fractionation of Multiple Cellular Membranes.J Virol. 2018 Mar 28;92(8):e01852-17. doi: 10.1128/JVI.01852-17. Print 2018 Apr 15. J Virol. 2018. PMID: 29367248 Free PMC article.
-
Poliovirus-induced changes in cellular membranes throughout infection.Curr Opin Virol. 2014 Dec;9:67-73. doi: 10.1016/j.coviro.2014.09.007. Epub 2014 Oct 11. Curr Opin Virol. 2014. PMID: 25310497 Free PMC article. Review.
-
Involvement of cellular membrane traffic proteins in poliovirus replication.Cell Cycle. 2007 Jan 1;6(1):36-8. doi: 10.4161/cc.6.1.3683. Epub 2007 Jan 11. Cell Cycle. 2007. PMID: 17245115 Review.
Cited by
-
Poliovirus infection transiently increases COPII vesicle budding.J Virol. 2012 Sep;86(18):9675-82. doi: 10.1128/JVI.01159-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740409 Free PMC article.
-
The Emerging Roles of Viroporins in ER Stress Response and Autophagy Induction during Virus Infection.Viruses. 2015 Jun 4;7(6):2834-57. doi: 10.3390/v7062749. Viruses. 2015. PMID: 26053926 Free PMC article. Review.
-
A novel, broad-spectrum inhibitor of enterovirus replication that targets host cell factor phosphatidylinositol 4-kinase IIIβ.Antimicrob Agents Chemother. 2013 Oct;57(10):4971-81. doi: 10.1128/AAC.01175-13. Epub 2013 Jul 29. Antimicrob Agents Chemother. 2013. PMID: 23896472 Free PMC article.
-
An integrated analysis of membrane remodeling during porcine reproductive and respiratory syndrome virus replication and assembly.PLoS One. 2018 Jul 24;13(7):e0200919. doi: 10.1371/journal.pone.0200919. eCollection 2018. PLoS One. 2018. PMID: 30040832 Free PMC article.
-
The Porcine Deltacoronavirus Replication Organelle Comprises Double-Membrane Vesicles and Zippered Endoplasmic Reticulum with Double-Membrane Spherules.Viruses. 2019 Nov 5;11(11):1030. doi: 10.3390/v11111030. Viruses. 2019. PMID: 31694296 Free PMC article.
References
-
- Agirre A, Barco A, Carrasco L, Nieva JL. 2002. Viroporin-mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J. Biol. Chem. 277:40434–40441 - PubMed
-
- Amako K, Dales S. 1967. Cytopathology of mengovirus infection. II. Proliferation of membranous cisternae. Virology 32:201–215 - PubMed
-
- Baumeister W, Grimm R, Walz J. 1999. Electron tomography of molecules and cells. Trends Cell Biol. 9:81–85 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

