FOG-1 and GATA-1 act sequentially to specify definitive megakaryocytic and erythroid progenitors
- PMID: 22068055
- PMCID: PMC3261555
- DOI: 10.1038/emboj.2011.390
FOG-1 and GATA-1 act sequentially to specify definitive megakaryocytic and erythroid progenitors
Abstract
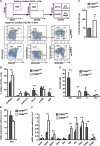
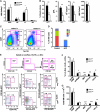
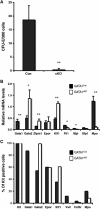
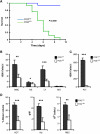
The transcription factors that control lineage specification of haematopoietic stem cells (HSCs) have been well described for the myeloid and lymphoid lineages, whereas transcriptional control of erythroid (E) and megakaryocytic (Mk) fate is less understood. We here use conditional removal of the GATA-1 and FOG-1 transcription factors to identify FOG-1 as required for the formation of all committed Mk- and E-lineage progenitors, whereas GATA-1 was observed to be specifically required for E-lineage commitment. FOG-1-deficient HSCs and preMegEs, the latter normally bipotent for the Mk and E lineages, underwent myeloid transcriptional reprogramming, and formed myeloid, but not erythroid and megakaryocytic cells in vitro. These results identify FOG-1 and GATA-1 as required for formation of bipotent Mk/E progenitors and their E-lineage commitment, respectively, and show that FOG-1 mediates transcriptional Mk/E programming of HSCs as well as their subsequent Mk/E-lineage commitment. Finally, C/EBPs and FOG-1 exhibited transcriptional cross-regulation in early myelo-erythroid progenitors making their functional antagonism a potential mechanism for separation of the myeloid and Mk/E lineages.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
FOG1 requires NuRD to promote hematopoiesis and maintain lineage fidelity within the megakaryocytic-erythroid compartment.Blood. 2010 Mar 18;115(11):2156-66. doi: 10.1182/blood-2009-10-251280. Epub 2010 Jan 11. Blood. 2010. PMID: 20065294 Free PMC article.
-
Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage.J Exp Med. 2008 Mar 17;205(3):611-24. doi: 10.1084/jem.20070544. Epub 2008 Feb 25. J Exp Med. 2008. PMID: 18299398 Free PMC article.
-
MEIS1 regulates early erythroid and megakaryocytic cell fate.Haematologica. 2014 Oct;99(10):1555-64. doi: 10.3324/haematol.2014.106567. Epub 2014 Aug 8. Haematologica. 2014. PMID: 25107888 Free PMC article.
-
A regulatory network governing Gata1 and Gata2 gene transcription orchestrates erythroid lineage differentiation.Int J Hematol. 2014 Nov;100(5):417-24. doi: 10.1007/s12185-014-1568-0. Epub 2014 Mar 18. Int J Hematol. 2014. PMID: 24638828 Review.
-
Concise Review: Bipotent Megakaryocytic-Erythroid Progenitors: Concepts and Controversies.Stem Cells. 2018 Aug;36(8):1138-1145. doi: 10.1002/stem.2834. Epub 2018 May 2. Stem Cells. 2018. PMID: 29658164 Free PMC article. Review.
Cited by
-
Pluripotent stem cells reveal erythroid-specific activities of the GATA1 N-terminus.J Clin Invest. 2015 Mar 2;125(3):993-1005. doi: 10.1172/JCI75714. Epub 2015 Jan 26. J Clin Invest. 2015. PMID: 25621499 Free PMC article.
-
Deciphering transcriptome alterations in bone marrow hematopoiesis at single-cell resolution in immune thrombocytopenia.Signal Transduct Target Ther. 2022 Oct 7;7(1):347. doi: 10.1038/s41392-022-01167-9. Signal Transduct Target Ther. 2022. PMID: 36202780 Free PMC article.
-
SPI1-KLF1/LYL1 axis regulates lineage commitment during endothelial-to-hematopoietic transition from human pluripotent stem cells.iScience. 2024 Jun 28;27(8):110409. doi: 10.1016/j.isci.2024.110409. eCollection 2024 Aug 16. iScience. 2024. PMID: 39108738 Free PMC article.
-
C/EBPα is required for long-term self-renewal and lineage priming of hematopoietic stem cells and for the maintenance of epigenetic configurations in multipotent progenitors.PLoS Genet. 2014 Jan;10(1):e1004079. doi: 10.1371/journal.pgen.1004079. Epub 2014 Jan 9. PLoS Genet. 2014. PMID: 24415956 Free PMC article.
-
Megakaryopoiesis and Platelet Biology: Roles of Transcription Factors and Emerging Clinical Implications.Int J Mol Sci. 2021 Sep 5;22(17):9615. doi: 10.3390/ijms22179615. Int J Mol Sci. 2021. PMID: 34502524 Free PMC article. Review.
References
-
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, Anderson K, Sitnicka E, Sasaki Y, Sigvardsson M, Jacobsen SE (2005) Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121: 295–306 - PubMed
-
- Akashi K, Traver D, Miyamoto T, Weissman IL (2000) A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404: 193–197 - PubMed
-
- Amigo JD, Ackermann GE, Cope JJ, Yu M, Cooney JD, Ma D, Langer NB, Shafizadeh E, Shaw GC, Horsely W, Trede NS, Davidson AJ, Barut BA, Zhou Y, Wojiski SA, Traver D, Moran TB, Kourkoulis G, Hsu K, Kanki JP et al. (2009) The role and regulation of friend of GATA-1 (FOG-1) during blood development in the zebrafish. Blood 114: 4654–4663 - PMC - PubMed
-
- Athanasiou M, Clausen PA, Mavrothalassitis GJ, Zhang XK, Watson DK, Blair DG (1996) Increased expression of the ETS-related transcription factor FLI-1/ERGB correlates with and can induce the megakaryocytic phenotype. Cell growth Differ 7: 1525–1534 - PubMed
-
- Athanasiou M, Mavrothalassitis G, Sun-Hoffman L, Blair DG (2000) FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia 14: 439–445 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

