S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance
- PMID: 22064703
- PMCID: PMC3357991
- DOI: 10.1038/cr.2011.176
S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance
Abstract
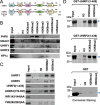
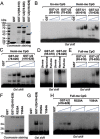
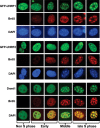
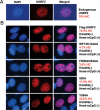
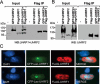
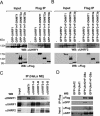
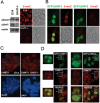
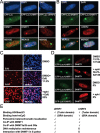
Recent studies demonstrate that UHRF1 is required for DNA methylation maintenance by targeting DNMT1 to DNA replication foci, presumably through its unique hemi-methylated DNA-binding activity and interaction with DNMT1. UHRF2, another member of the UHRF family proteins, is highly similar to UHRF1 in both sequence and structure, raising questions about its role in DNA methylation. In this study, we demonstrate that, like UHRF1, UHRF2 also binds preferentially to methylated histone H3 lysine 9 (H3K9) through its conserved tudor domain and hemi-methylated DNA through the SET and Ring associated domain. Like UHRF1, UHRF2 is enriched in pericentric heterochromatin. The heterochromatin localization depends to large extent on its methylated H3K9-binding activity and to less extent on its methylated DNA-binding activity. Coimmunoprecipitation experiments demonstrate that both UHRF1 and UHRF2 interact with DNMT1, DNMT3a, DNMT3b and G9a. Despite all these conserved functions, we find that UHRF2 is not able to rescue the DNA methylation defect in Uhrf1 null mouse embryonic stem cells. This can be attributed to the inability for UHRF2 to recruit DNMT1 to replication foci during S phase of the cell cycle. Indeed, we find that while UHRF1 interacts with DNMT1 in an S phase-dependent manner in cells, UHRF2 does not. Thus, our study demonstrates that UHRF2 and UHRF1 are not functionally redundant in DNA methylation maintenance and reveals the cell-cycle-dependent interaction between UHRF1 and DNMT1 as a key regulatory mechanism targeting DNMT1 for DNA methylation.
Figures
Similar articles
-
UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9.Nat Commun. 2013;4:1563. doi: 10.1038/ncomms2562. Nat Commun. 2013. PMID: 23463006
-
Comparative biochemical analysis of UHRF proteins reveals molecular mechanisms that uncouple UHRF2 from DNA methylation maintenance.Nucleic Acids Res. 2018 May 18;46(9):4405-4416. doi: 10.1093/nar/gky151. Nucleic Acids Res. 2018. PMID: 29506131 Free PMC article.
-
The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA.J Biol Chem. 2014 Jan 3;289(1):379-86. doi: 10.1074/jbc.M113.523209. Epub 2013 Nov 19. J Biol Chem. 2014. PMID: 24253042 Free PMC article.
-
Coordinated Dialogue between UHRF1 and DNMT1 to Ensure Faithful Inheritance of Methylated DNA Patterns.Genes (Basel). 2019 Jan 18;10(1):65. doi: 10.3390/genes10010065. Genes (Basel). 2019. PMID: 30669400 Free PMC article. Review.
-
Regulatory mechanism and biological function of UHRF1-DNMT1-mediated DNA methylation.Funct Integr Genomics. 2022 Dec;22(6):1113-1126. doi: 10.1007/s10142-022-00918-9. Epub 2022 Nov 14. Funct Integr Genomics. 2022. PMID: 36372834 Review.
Cited by
-
The nuclear protein UHRF2 is a direct target of the transcription factor E2F1 in the induction of apoptosis.J Biol Chem. 2013 Aug 16;288(33):23833-43. doi: 10.1074/jbc.M112.447276. Epub 2013 Jul 5. J Biol Chem. 2013. PMID: 23833190 Free PMC article.
-
UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9.Nat Commun. 2013;4:1563. doi: 10.1038/ncomms2562. Nat Commun. 2013. PMID: 23463006
-
Identification of UHRF2 as a novel DNA interstrand crosslink sensor protein.PLoS Genet. 2018 Oct 18;14(10):e1007643. doi: 10.1371/journal.pgen.1007643. eCollection 2018 Oct. PLoS Genet. 2018. PMID: 30335751 Free PMC article.
-
Association of UHRF1 gene polymorphisms with oligospermia in Chinese males.J Assist Reprod Genet. 2019 Dec;36(12):2563-2573. doi: 10.1007/s10815-019-01614-7. Epub 2019 Dec 4. J Assist Reprod Genet. 2019. PMID: 31802345 Free PMC article.
-
Comparative biochemical analysis of UHRF proteins reveals molecular mechanisms that uncouple UHRF2 from DNA methylation maintenance.Nucleic Acids Res. 2018 May 18;46(9):4405-4416. doi: 10.1093/nar/gky151. Nucleic Acids Res. 2018. PMID: 29506131 Free PMC article.
References
-
- Taberlay PC, Jones PA. DNA methylation and cancer. Prog Drug Res. 2011;67:1–23. - PubMed
-
- Chen T, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Bio. 2004;60:55–89. - PubMed
-
- Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

