DNA glycosylases: in DNA repair and beyond
- PMID: 22048164
- PMCID: PMC3260424
- DOI: 10.1007/s00412-011-0347-4
DNA glycosylases: in DNA repair and beyond
Abstract
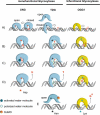
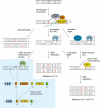
The base excision repair machinery protects DNA in cells from the damaging effects of oxidation, alkylation, and deamination; it is specialized to fix single-base damage in the form of small chemical modifications. Base modifications can be mutagenic and/or cytotoxic, depending on how they interfere with the template function of the DNA during replication and transcription. DNA glycosylases play a key role in the elimination of such DNA lesions; they recognize and excise damaged bases, thereby initiating a repair process that restores the regular DNA structure with high accuracy. All glycosylases share a common mode of action for damage recognition; they flip bases out of the DNA helix into a selective active site pocket, the architecture of which permits a sensitive detection of even minor base irregularities. Within the past few years, it has become clear that nature has exploited this ability to read the chemical structure of DNA bases for purposes other than canonical DNA repair. DNA glycosylases have been brought into context with molecular processes relating to innate and adaptive immunity as well as to the control of DNA methylation and epigenetic stability. Here, we summarize the key structural and mechanistic features of DNA glycosylases with a special focus on the mammalian enzymes, and then review the evidence for the newly emerging biological functions beyond the protection of genome integrity.
Figures



Similar articles
-
Emerging Roles of DNA Glycosylases and the Base Excision Repair Pathway.Trends Biochem Sci. 2019 Sep;44(9):765-781. doi: 10.1016/j.tibs.2019.04.006. Epub 2019 May 9. Trends Biochem Sci. 2019. PMID: 31078398 Free PMC article. Review.
-
Oxidative DNA damage repair in mammalian cells: a new perspective.DNA Repair (Amst). 2007 Apr 1;6(4):470-80. doi: 10.1016/j.dnarep.2006.10.011. Epub 2006 Nov 20. DNA Repair (Amst). 2007. PMID: 17116430 Free PMC article. Review.
-
Automated AFM analysis of DNA bending reveals initial lesion sensing strategies of DNA glycosylases.Sci Rep. 2020 Sep 23;10(1):15484. doi: 10.1038/s41598-020-72102-7. Sci Rep. 2020. PMID: 32968112 Free PMC article.
-
Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2.DNA Repair (Amst). 2005 Jan 2;4(1):41-50. doi: 10.1016/j.dnarep.2004.07.006. DNA Repair (Amst). 2005. PMID: 15533836
-
DNA repair in mammalian cells: Base excision repair: the long and short of it.Cell Mol Life Sci. 2009 Mar;66(6):981-93. doi: 10.1007/s00018-009-8736-z. Cell Mol Life Sci. 2009. PMID: 19153658 Free PMC article. Review.
Cited by
-
Targeting the DNA damage response in cancer.MedComm (2020). 2024 Oct 31;5(11):e788. doi: 10.1002/mco2.788. eCollection 2024 Nov. MedComm (2020). 2024. PMID: 39492835 Free PMC article. Review.
-
Complex DNA repair pathways as possible therapeutic targets to overcome temozolomide resistance in glioblastoma.Front Oncol. 2012 Dec 5;2:186. doi: 10.3389/fonc.2012.00186. eCollection 2012. Front Oncol. 2012. PMID: 23227453 Free PMC article.
-
Expanding molecular roles of UV-DDB: Shining light on genome stability and cancer.DNA Repair (Amst). 2020 Oct;94:102860. doi: 10.1016/j.dnarep.2020.102860. Epub 2020 Apr 27. DNA Repair (Amst). 2020. PMID: 32739133 Free PMC article. Review.
-
MutS-Homolog2 silencing generates tetraploid meiocytes in tomato (Solanum lycopersicum).Plant Direct. 2018 Jan 2;2(1):e00017. doi: 10.1002/pld3.17. eCollection 2018 Jan. Plant Direct. 2018. PMID: 31245679 Free PMC article.
-
XRCC1 Prevents Replication Fork Instability during Misincorporation of the DNA Demethylation Bases 5-Hydroxymethyl-2'-Deoxycytidine and 5-Hydroxymethyl-2'-Deoxyuridine.Int J Mol Sci. 2022 Jan 14;23(2):893. doi: 10.3390/ijms23020893. Int J Mol Sci. 2022. PMID: 35055077 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

