Identifying polyglutamine protein species in situ that best predict neurodegeneration
- PMID: 22037470
- PMCID: PMC3271120
- DOI: 10.1038/nchembio.694
Identifying polyglutamine protein species in situ that best predict neurodegeneration
Erratum in
- Nat Chem Biol. 2012 Mar;8(3):318
Abstract
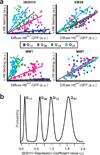
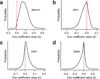
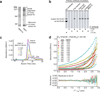
Polyglutamine (polyQ) stretches exceeding a threshold length confer a toxic function to proteins that contain them and cause at least nine neurological disorders. The basis for this toxicity threshold is unclear. Although polyQ expansions render proteins prone to aggregate into inclusion bodies, this may be a neuronal coping response to more toxic forms of polyQ. The exact structure of these more toxic forms is unknown. Here we show that the monoclonal antibody 3B5H10 recognizes a species of polyQ protein in situ that strongly predicts neuronal death. The epitope selectively appears among some of the many low-molecular-weight conformational states assumed by expanded polyQ and disappears in higher-molecular-weight aggregated forms, such as inclusion bodies. These results suggest that protein monomers and possibly small oligomers containing expanded polyQ stretches can adopt a conformation that is recognized by 3B5H10 and is toxic or closely related to a toxic species.
Figures

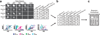


Comment in
-
Protein misfolding: Tracking a toxic polyQ epitope.Nat Chem Biol. 2011 Nov 15;7(12):861-2. doi: 10.1038/nchembio.718. Nat Chem Biol. 2011. PMID: 22086286 No abstract available.
-
A handle on neurodegenerative disease complexity.Nat Methods. 2012 Jan;9(1):21. doi: 10.1038/nmeth.1847. Nat Methods. 2012. PMID: 22312633 No abstract available.
Similar articles
-
Linear and extended: a common polyglutamine conformation recognized by the three antibodies MW1, 1C2 and 3B5H10.Hum Mol Genet. 2013 Oct 15;22(20):4215-23. doi: 10.1093/hmg/ddt273. Epub 2013 Jun 17. Hum Mol Genet. 2013. PMID: 23777629
-
Anti-PolyQ Antibodies Recognize a Short PolyQ Stretch in Both Normal and Mutant Huntingtin Exon 1.J Mol Biol. 2015 Jul 31;427(15):2507-2519. doi: 10.1016/j.jmb.2015.05.023. Epub 2015 Jun 3. J Mol Biol. 2015. PMID: 26047735 Free PMC article.
-
AQAMAN, a bisamidine-based inhibitor of toxic protein inclusions in neurons, ameliorates cytotoxicity in polyglutamine disease models.J Biol Chem. 2019 Feb 22;294(8):2757-2770. doi: 10.1074/jbc.RA118.006307. Epub 2018 Dec 28. J Biol Chem. 2019. PMID: 30593503 Free PMC article.
-
PolyQ disease: misfiring of a developmental cell death program?Trends Cell Biol. 2013 Apr;23(4):168-74. doi: 10.1016/j.tcb.2012.11.003. Epub 2012 Dec 8. Trends Cell Biol. 2013. PMID: 23228508 Free PMC article. Review.
-
Proteins Containing Expanded Polyglutamine Tracts and Neurodegenerative Disease.Biochemistry. 2017 Mar 7;56(9):1199-1217. doi: 10.1021/acs.biochem.6b00936. Epub 2017 Feb 21. Biochemistry. 2017. PMID: 28170216 Free PMC article. Review.
Cited by
-
Spatial sequestration and detoxification of Huntingtin by the ribosome quality control complex.Elife. 2016 Apr 1;5:e11792. doi: 10.7554/eLife.11792. Elife. 2016. PMID: 27033550 Free PMC article.
-
Native mutant huntingtin in human brain: evidence for prevalence of full-length monomer.J Biol Chem. 2012 Apr 13;287(16):13487-99. doi: 10.1074/jbc.M111.286609. Epub 2012 Feb 27. J Biol Chem. 2012. PMID: 22375012 Free PMC article.
-
Proteasome-mediated proteolysis of the polyglutamine-expanded androgen receptor is a late event in spinal and bulbar muscular atrophy (SBMA) pathogenesis.J Biol Chem. 2015 May 15;290(20):12572-84. doi: 10.1074/jbc.M114.617894. Epub 2015 Mar 20. J Biol Chem. 2015. PMID: 25795778 Free PMC article.
-
From Pathogenesis to Therapeutics: A Review of 150 Years of Huntington's Disease Research.Int J Mol Sci. 2023 Aug 21;24(16):13021. doi: 10.3390/ijms241613021. Int J Mol Sci. 2023. PMID: 37629202 Free PMC article. Review.
-
Amyloid modifier SERF1a interacts with polyQ-expanded huntingtin-exon 1 via helical interactions and exacerbates polyQ-induced toxicity.Commun Biol. 2023 Jul 21;6(1):767. doi: 10.1038/s42003-023-05142-0. Commun Biol. 2023. PMID: 37479809 Free PMC article.
References
-
- THDCR G. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. - PubMed
-
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. - PubMed
-
- Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. - PubMed
-
- Ko J, Ou S, Patterson PH. New anti-huntingtin monoclonal antibodies: Implications for huntingtin conformation and its binding proteins. Brain Res. Bull. 2001;56:319–329. - PubMed
-
- Rakhit R, et al. An immunological epitope selective for pathological monomer-misfolded SOD1 in ALS. Nat. Med. 2007;13:754–759. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

