The story so far: post-translational regulation of peroxisome proliferator-activated receptors by ubiquitination and SUMOylation
- PMID: 22037188
- PMCID: PMC3353776
- DOI: 10.1152/ajpheart.00703.2011
The story so far: post-translational regulation of peroxisome proliferator-activated receptors by ubiquitination and SUMOylation
Erratum in
- Am J Physiol Heart Circ Physiol. 2014 Aug 1;307(3):H464-6
Abstract
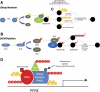
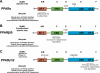
Many studies have implicated the peroxisome proliferator-activated receptor (PPAR) family of nuclear receptor transcription factors in regulating cardiac substrate metabolism and ATP generation. Recently, evidence from a variety of cell culture and organ systems has implicated ubiquitin and small ubiquitin-like modifier (SUMO) conjugation as post-translational modifications that regulate the activity of PPAR transcription factors and their coreceptors/coactivators. Here we introduce the ubiquitin and SUMO conjugation systems and extensively review how they have been shown to regulate all three PPAR isoforms (PPARα, PPARβ/δ, and PPARγ) in addition to the retinoid X receptor and PPARγ coactivator-1α subunits of the larger PPAR transcription factor complex. We then present how the specific ubiquitin (E3) ligases have been implicated and review emerging evidence that post-translational modifications of PPARs with ubiquitin and/or SUMO may play a role in cardiac disease. Because PPAR activity is perturbed in a variety of forms of heart disease and specific proteins regulate this process (E3 ligases), this may be a fruitful area of investigation with respect to finding new therapeutic targets.
Figures
Similar articles
-
Functional Regulation of PPARs through Post-Translational Modifications.Int J Mol Sci. 2018 Jun 12;19(6):1738. doi: 10.3390/ijms19061738. Int J Mol Sci. 2018. PMID: 29895749 Free PMC article. Review.
-
Peroxisome proliferator-activated receptor (PPAR)β/δ, a possible nexus of PPARα- and PPARγ-dependent molecular pathways in neurodegenerative diseases: Review and novel hypotheses.Neurochem Int. 2013 Oct;63(4):322-30. doi: 10.1016/j.neuint.2013.06.012. Epub 2013 Jun 25. Neurochem Int. 2013. PMID: 23811400 Review.
-
PPAR SUMOylation: some useful experimental tips.Methods Mol Biol. 2013;952:145-61. doi: 10.1007/978-1-62703-155-4_10. Methods Mol Biol. 2013. PMID: 23100230
-
PPARs and Myocardial Infarction.Int J Mol Sci. 2020 Dec 11;21(24):9436. doi: 10.3390/ijms21249436. Int J Mol Sci. 2020. PMID: 33322384 Free PMC article. Review.
-
Peroxisome proliferator-activated receptor (PPAR)-gamma positively controls and PPARalpha negatively controls cyclooxygenase-2 expression in rat brain astrocytes through a convergence on PPARbeta/delta via mutual control of PPAR expression levels.Mol Pharmacol. 2009 Aug;76(2):414-24. doi: 10.1124/mol.109.056010. Epub 2009 May 29. Mol Pharmacol. 2009. PMID: 19483106
Cited by
-
Not So Slim Anymore-Evidence for the Role of SUMO in the Regulation of Lipid Metabolism.Biomolecules. 2020 Aug 6;10(8):1154. doi: 10.3390/biom10081154. Biomolecules. 2020. PMID: 32781719 Free PMC article. Review.
-
PPARgamma-Dependent Control of Renin Expression: Molecular Mechanisms and Pathophysiological Relevance.PPAR Res. 2013;2013:451016. doi: 10.1155/2013/451016. Epub 2013 Oct 30. PPAR Res. 2013. PMID: 24288524 Free PMC article. Review.
-
Hepatic PPARα function is controlled by polyubiquitination and proteasome-mediated degradation through the coordinated actions of PAQR3 and HUWE1.Hepatology. 2018 Jul;68(1):289-303. doi: 10.1002/hep.29786. Epub 2018 Apr 26. Hepatology. 2018. PMID: 29331071 Free PMC article.
-
SUMOylation as a Therapeutic Target for Myocardial Infarction.Front Cardiovasc Med. 2021 Jul 28;8:701583. doi: 10.3389/fcvm.2021.701583. eCollection 2021. Front Cardiovasc Med. 2021. PMID: 34395563 Free PMC article. Review.
-
The mito-DAMP cardiolipin blocks IL-10 production causing persistent inflammation during bacterial pneumonia.Nat Commun. 2017 Jan 11;8:13944. doi: 10.1038/ncomms13944. Nat Commun. 2017. PMID: 28074841 Free PMC article.
References
-
- Adhihetty PJ, Uguccioni G, Leick L, Hidalgo J, Pilegaard H, Hood DA. The role of PGC-1alpha on mitochondrial function and apoptotic susceptibility in muscle. Am J Physiol Cell Physiol 297: C217–C225, 2009 - PubMed
-
- Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, Rybkin II, Shelton JM, Manieri M, Cinti S, Schoen FJ, Bassel-Duby R, Rosenzweig A, Ingwall JS, Spiegelman BM. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab 1: 259–271, 2005 - PubMed
-
- Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J 16: 1879–1886, 2002 - PubMed
-
- Bao Y, Li R, Jiang J, Cai B, Gao J, Le K, Zhang F, Chen S, Liu P. Activation of peroxisome proliferator-activated receptor gamma inhibits endothelin-1-induced cardiac hypertrophy via the calcineurin/NFAT signaling pathway. Mol Cell Biochem 317: 189–196, 2008 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

