Master transcription factors determine cell-type-specific responses to TGF-β signaling
- PMID: 22036565
- PMCID: PMC3212730
- DOI: 10.1016/j.cell.2011.08.050
Master transcription factors determine cell-type-specific responses to TGF-β signaling
Abstract
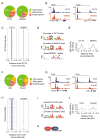
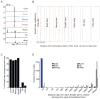
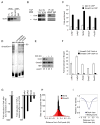
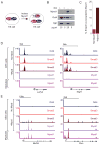
Transforming growth factor beta (TGF-β) signaling, mediated through the transcription factors Smad2 and Smad3 (Smad2/3), directs different responses in different cell types. Here we report that Smad3 co-occupies the genome with cell-type-specific master transcription factors. Thus, Smad3 occupies the genome with Oct4 in embryonic stem cells (ESCs), Myod1 in myotubes, and PU.1 in pro-B cells. We find that these master transcription factors are required for Smad3 occupancy and that TGF-β signaling largely affects the genes bound by the master transcription factors. Furthermore, we show that induction of Myod1 in nonmuscle cells is sufficient to redirect Smad3 to Myod1 sites. We conclude that cell-type-specific master transcription factors determine the genes bound by Smad2/3 and are thus responsible for orchestrating the cell-type-specific effects of TGF-β signaling.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells.Cell Rep. 2013 Dec 26;5(6):1611-24. doi: 10.1016/j.celrep.2013.11.021. Epub 2013 Dec 12. Cell Rep. 2013. PMID: 24332857
-
A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling.J Cell Biochem. 2007 May 1;101(1):9-33. doi: 10.1002/jcb.21255. J Cell Biochem. 2007. PMID: 17340614 Review.
-
Functional characterization of transforming growth factor beta signaling in Smad2- and Smad3-deficient fibroblasts.J Biol Chem. 2001 Jun 8;276(23):19945-53. doi: 10.1074/jbc.M102382200. Epub 2001 Mar 21. J Biol Chem. 2001. PMID: 11262418
-
TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3.Genes Dev. 2001 Nov 15;15(22):2950-66. doi: 10.1101/gad.925901. Genes Dev. 2001. PMID: 11711431 Free PMC article.
-
TGF-β control of stem cell differentiation genes.FEBS Lett. 2012 Jul 4;586(14):1953-8. doi: 10.1016/j.febslet.2012.03.023. Epub 2012 Apr 10. FEBS Lett. 2012. PMID: 22710171 Free PMC article. Review.
Cited by
-
Transcriptional mechanisms of pancreatic β-cell maturation and functional adaptation.Trends Endocrinol Metab. 2021 Jul;32(7):474-487. doi: 10.1016/j.tem.2021.04.011. Epub 2021 May 21. Trends Endocrinol Metab. 2021. PMID: 34030925 Free PMC article. Review.
-
Human genetic variation within neural crest enhancers: molecular and phenotypic implications.Philos Trans R Soc Lond B Biol Sci. 2013 May 6;368(1620):20120360. doi: 10.1098/rstb.2012.0360. Print 2013. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23650634 Free PMC article. Review.
-
CNS repair and axon regeneration: Using genetic variation to determine mechanisms.Exp Neurol. 2017 Jan;287(Pt 3):409-422. doi: 10.1016/j.expneurol.2016.05.004. Epub 2016 May 6. Exp Neurol. 2017. PMID: 27163547 Free PMC article. Review.
-
Transcription Factors in Cartilage Homeostasis and Osteoarthritis.Biology (Basel). 2020 Sep 14;9(9):290. doi: 10.3390/biology9090290. Biology (Basel). 2020. PMID: 32937960 Free PMC article. Review.
-
Phospho-specific Smad3 signaling: impact on breast oncogenesis.Cell Cycle. 2012 Jul 1;11(13):2443-51. doi: 10.4161/cc.20546. Epub 2012 Jul 1. Cell Cycle. 2012. PMID: 22659843 Free PMC article. Review.
References
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
-
- Beattie GM, Lopez AD, Bucay N, Hinton A, Firpo MT, King CC, Hayek A. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells. 2005;23:489–495. - PubMed
-
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. - PubMed
-
- Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110:19–32. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

