TR-FRET-based high-throughput screening assay for identification of UBC13 inhibitors
- PMID: 22034497
- PMCID: PMC4172584
- DOI: 10.1177/1087057111423417
TR-FRET-based high-throughput screening assay for identification of UBC13 inhibitors
Abstract
UBC13 is a noncanonical ubiquitin conjugating enzyme (E2) that has been implicated in a variety of cellular signaling processes due to its ability to catalyze formation of lysine 63-linked polyubiquitin chains on various substrates. In particular, UBC13 is required for signaling by a variety of receptors important in immune regulation, making it a candidate target for inflammatory diseases. UBC13 is also critical for double-strand DNA repair and thus a potential radiosensitizer and chemosensitizer target for oncology. The authors developed a high-throughput screening (HTS) assay for UBC13 based on the method of time-resolved fluorescence resonance energy transfer (TR-FRET). The TR-FRET assay combines fluorochrome (Fl)-conjugated ubiquitin (fluorescence acceptor) with terbium (Tb)-conjugated ubiquitin (fluorescence donor), such that the assembly of mixed chains of Fl- and Tb-ubiquitin creates a robust TR-FRET signal. The authors defined conditions for optimized performance of the TR-FRET assay in both 384- and 1536-well formats. Chemical library screens (total 456 865 compounds) were conducted in high-throughput mode using various compound collections, affording superb Z' scores (typically >0.7) and thus validating the performance of the assays. Altogether, the HTS assays described here are suitable for large-scale, automated screening of chemical libraries in search of compounds with inhibitory activity against UBC13.
Figures

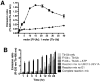


 : Full inhibition controls include reaction system lacking UBC13-UEV1A without compound added.
: Full inhibition controls include reaction system lacking UBC13-UEV1A without compound added.  : No inhibition controls include complete reaction system without compound added.
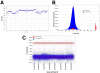
: No inhibition controls include complete reaction system without compound added.  : LOPAC compounds; x-axis: Well #; y-axis: % Inhibition; z-axis: Plate ID.
: LOPAC compounds; x-axis: Well #; y-axis: % Inhibition; z-axis: Plate ID.

Similar articles
-
Development of a High-Throughput Assay for Inhibitors of the Polo-Box Domain of Polo-Like Kinase 1 Based on Time-Resolved Fluorescence Energy Transfer.Biol Pharm Bull. 2017;40(9):1454-1462. doi: 10.1248/bpb.b17-00283. Biol Pharm Bull. 2017. PMID: 28867728
-
A High-Throughput Screening Strategy for Development of RNF8-Ubc13 Protein-Protein Interaction Inhibitors.SLAS Discov. 2017 Mar;22(3):316-323. doi: 10.1177/1087057116681408. Epub 2016 Dec 13. SLAS Discov. 2017. PMID: 27909234
-
Development of a high-throughput assay to identify inhibitors of the ubiquitin-conjugating enzyme UBCH10.SLAS Discov. 2022 Jun;27(4):266-271. doi: 10.1016/j.slasd.2022.03.007. Epub 2022 Mar 25. SLAS Discov. 2022. PMID: 35342035
-
Ubc13: the Lys63 ubiquitin chain building machine.Oncotarget. 2016 Sep 27;7(39):64471-64504. doi: 10.18632/oncotarget.10948. Oncotarget. 2016. PMID: 27486774 Free PMC article. Review.
-
Selective UBC 13 Inhibitors.2012 Apr 16 [updated 2014 May 13]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010–. 2012 Apr 16 [updated 2014 May 13]. In: Probe Reports from the NIH Molecular Libraries Program [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010–. PMID: 23762936 Free Books & Documents. Review.
Cited by
-
A novel small-molecule inhibitor of influenza A virus acts by suppressing PA endonuclease activity of the viral polymerase.Sci Rep. 2016 Mar 9;6:22880. doi: 10.1038/srep22880. Sci Rep. 2016. PMID: 26956222 Free PMC article.
-
Targeting TRIM Proteins: A Quest towards Drugging an Emerging Protein Class.Chembiochem. 2021 Jun 15;22(12):2011-2031. doi: 10.1002/cbic.202000787. Epub 2021 Mar 18. Chembiochem. 2021. PMID: 33482040 Free PMC article. Review.
-
Application of FRET Biosensors in Mechanobiology and Mechanopharmacological Screening.Front Bioeng Biotechnol. 2020 Nov 9;8:595497. doi: 10.3389/fbioe.2020.595497. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33240867 Free PMC article. Review.
-
Inhibition of proliferation and survival of diffuse large B-cell lymphoma cells by a small-molecule inhibitor of the ubiquitin-conjugating enzyme Ubc13-Uev1A.Blood. 2012 Aug 23;120(8):1668-77. doi: 10.1182/blood-2012-02-406074. Epub 2012 Jul 12. Blood. 2012. PMID: 22791293 Free PMC article.
-
Polypharmacology of small molecules targeting the ubiquitin-proteasome and ubiquitin-like systems.Oncotarget. 2015;6(12):9646-56. doi: 10.18632/oncotarget.3917. Oncotarget. 2015. PMID: 25991664 Free PMC article. Review.
References
-
- Boisclair MD, McClure C, Josiah S, Glass S, Bottomley S, Kamerkar S, Hemmila I. Development of a ubiquitin transfer assay for high throughput screening by fluorescence resonance energy transfer. J Biomol Screen. 2000;5:319–328. - PubMed
-
- Carlson CB, Horton RA, Vogel KW. A Toolbox Approach to High-Throughput TR-FRET-Based SUMOylation and DeSUMOylation Assays. Assay Drug Dev Technol. 2009 - PubMed
-
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. - PubMed
-
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

