Identical functional organization of nonpolytene and polytene chromosomes in Drosophila melanogaster
- PMID: 22022482
- PMCID: PMC3191165
- DOI: 10.1371/journal.pone.0025960
Identical functional organization of nonpolytene and polytene chromosomes in Drosophila melanogaster
Erratum in
- PLoS One. 2011;6(11). doi:10.1371/annotation/45b44e2a-c751-418b-bbb7-7023998abdfc
Abstract
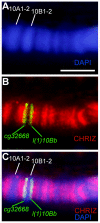
Salivary gland polytene chromosomes demonstrate banding pattern, genetic meaning of which is an enigma for decades. Till now it is not known how to mark the band/interband borders on physical map of DNA and structures of polytene chromosomes are not characterized in molecular and genetic terms. It is not known either similar banding pattern exists in chromosomes of regular diploid mitotically dividing nonpolytene cells. Using the newly developed approach permitting to identify the interband material and localization data of interband-specific proteins from modENCODE and other genome-wide projects, we identify physical limits of bands and interbands in small cytological region 9F13-10B3 of the X chromosome in D. melanogaster, as well as characterize their general molecular features. Our results suggests that the polytene and interphase cell line chromosomes have practically the same patterns of bands and interbands reflecting, probably, the basic principle of interphase chromosome organization. Two types of bands have been described in chromosomes, early and late-replicating, which differ in many aspects of their protein and genetic content. As appeared, origin recognition complexes are located almost totally in the interbands of chromosomes.
Conflict of interest statement
Figures

Similar articles
-
Genetic organization of interphase chromosome bands and interbands in Drosophila melanogaster.PLoS One. 2014 Jul 29;9(7):e101631. doi: 10.1371/journal.pone.0101631. eCollection 2014. PLoS One. 2014. PMID: 25072930 Free PMC article.
-
[Chromomeric organization of interphase chromosomes in Drosophila melanogaster].Tsitologiia. 2013;55(3):144-7. Tsitologiia. 2013. PMID: 23795454 Review. Russian.
-
Banding patterns in Drosophila melanogaster polytene chromosomes correlate with DNA-binding protein occupancy.Bioessays. 2012 Jun;34(6):498-508. doi: 10.1002/bies.201100142. Epub 2012 Mar 15. Bioessays. 2012. PMID: 22419120 Review.
-
Similarity in replication timing between polytene and diploid cells is associated with the organization of the Drosophila genome.PLoS One. 2018 Apr 16;13(4):e0195207. doi: 10.1371/journal.pone.0195207. eCollection 2018. PLoS One. 2018. PMID: 29659604 Free PMC article.
-
Genes Containing Long Introns Occupy Series of Bands and Interbands In Drosophila melanogaster polytene Chromosomes.Genes (Basel). 2020 Apr 11;11(4):417. doi: 10.3390/genes11040417. Genes (Basel). 2020. PMID: 32290448 Free PMC article.
Cited by
-
Mechanism and functional role of the interaction between CP190 and the architectural protein Pita in Drosophila melanogaster.Epigenetics Chromatin. 2021 Mar 22;14(1):16. doi: 10.1186/s13072-021-00391-x. Epigenetics Chromatin. 2021. PMID: 33752739 Free PMC article.
-
Super-resolution microscopy reveals stochastic initiation of replication in Drosophila polytene chromosomes.Chromosome Res. 2022 Dec;30(4):361-383. doi: 10.1007/s10577-021-09679-w. Epub 2022 Feb 28. Chromosome Res. 2022. PMID: 35226231 Free PMC article.
-
Polytene Chromosomes - A Portrait of Functional Organization of the Drosophila Genome.Curr Genomics. 2018 Apr;19(3):179-191. doi: 10.2174/1389202918666171016123830. Curr Genomics. 2018. PMID: 29606905 Free PMC article. Review.
-
Genetic organization of interphase chromosome bands and interbands in Drosophila melanogaster.PLoS One. 2014 Jul 29;9(7):e101631. doi: 10.1371/journal.pone.0101631. eCollection 2014. PLoS One. 2014. PMID: 25072930 Free PMC article.
-
Quantified effects of chromosome-nuclear envelope attachments on 3D organization of chromosomes.Nucleus. 2015;6(3):212-24. doi: 10.1080/19491034.2015.1056441. Nucleus. 2015. PMID: 26068134 Free PMC article.
References
-
- Beermann W. Chromomeres and genes. In: Beermann W, editor. Results and problems in cell differentiation. V. 4. Berlin, Heidelberg, New York: Springer; 1972. pp. 1–33. - PubMed
-
- Zhimulev IF. Morphology and structure of polytene chromosomes. Adv Genet. 1996;34:1–497. - PubMed
-
- Zhimulev IF, Belyaeva ES, Semeshin VF, Koryakov DE, Demakov SA, et al. Polytene Chromosomes: 70 Years of Genetic Research. Int Rev Cytol. 2004;241:203–275. - PubMed
-
- Andreyenkova NG, Kokoza EB, Semeshin VF, Belyaeva ES, Demakov SA, et al. Localization and characteristics of DNA underreplication zone in the 75 C region of intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma. 2009;118:747–761. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

