PCBP2 enhances the antiviral activity of IFN-α against HCV by stabilizing the mRNA of STAT1 and STAT2
- PMID: 22022391
- PMCID: PMC3191149
- DOI: 10.1371/journal.pone.0025419
PCBP2 enhances the antiviral activity of IFN-α against HCV by stabilizing the mRNA of STAT1 and STAT2
Abstract
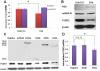
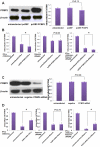
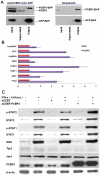
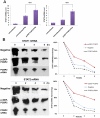
Interferon-α (IFN-α) is a natural choice for the treatment of hepatitis C, but half of the chronically infected individuals do not achieve sustained clearance of hepatitis C virus (HCV) during treatment with IFN-α alone. The virus can impair IFN-α signaling and cellular factors that have an effect on the viral life cycles. We found that the protein PCBP2 is down-regulated in HCV-replicon containing cells (R1b). However, the effects and mechanisms of PCBP2 on HCV are unclear. To determine the effect of PCBP2 on HCV, overexpression and knockdown of PCBP2 were performed in R1b cells. Interestingly, we found that PCBP2 can facilitate the antiviral activity of IFN-α against HCV, although the RNA level of HCV was unaffected by either the overexpression or absence of PCBP2 in R1b cells. RIP-qRT-PCR and RNA half-life further revealed that PCBP2 stabilizes the mRNA of STAT1 and STAT2 through binding the 3'Untranslated Region (UTR) of these two molecules, which are pivotal for the IFN-α anti-HCV effect. RNA pull-down assay confirmed that there were binding sites located in the C-rich tracts in the 3'UTR of their mRNAs. Stabilization of mRNA by PCBP2 leads to the increased protein expression of STAT1 and STAT2 and a consistent increase of phosphorylated STAT1 and STAT2. These effects, in turn, enhance the antiviral effect of IFN-α. These findings indicate that PCBP2 may play an important role in the IFN-α response against HCV and may benefit the HCV clinical therapy.
Conflict of interest statement
Figures


Similar articles
-
Inhibition of STAT Pathway Impairs Anti-Hepatitis C Virus Effect of Interferon Alpha.Cell Physiol Biochem. 2016;40(1-2):77-90. doi: 10.1159/000452526. Epub 2016 Nov 18. Cell Physiol Biochem. 2016. PMID: 27855377
-
Unphosphorylated ISGF3 drives constitutive expression of interferon-stimulated genes to protect against viral infections.Sci Signal. 2017 Apr 25;10(476):eaah4248. doi: 10.1126/scisignal.aah4248. Sci Signal. 2017. PMID: 28442624
-
STAT1 is essential for the inhibition of hepatitis C virus replication by interferon-λ but not by interferon-α.Sci Rep. 2016 Dec 8;6:38336. doi: 10.1038/srep38336. Sci Rep. 2016. PMID: 27929099 Free PMC article.
-
Understanding the molecular mechanism(s) of hepatitis C virus (HCV) induced interferon resistance.Infect Genet Evol. 2013 Oct;19:113-9. doi: 10.1016/j.meegid.2013.06.025. Epub 2013 Jul 5. Infect Genet Evol. 2013. PMID: 23831932 Review.
-
Mechanisms of Viral Degradation of Cellular Signal Transducer and Activator of Transcription 2.Int J Mol Sci. 2022 Jan 1;23(1):489. doi: 10.3390/ijms23010489. Int J Mol Sci. 2022. PMID: 35008916 Free PMC article. Review.
Cited by
-
Cellular STAT3 functions via PCBP2 to restrain Epstein-Barr Virus lytic activation in B lymphocytes.J Virol. 2015 May;89(9):5002-11. doi: 10.1128/JVI.00121-15. Epub 2015 Feb 25. J Virol. 2015. PMID: 25717101 Free PMC article.
-
Longitudinal study of surrogate aging measures during human immunodeficiency virus seroconversion.Aging (Albany NY). 2017 Feb 23;9(3):687-705. doi: 10.18632/aging.101184. Aging (Albany NY). 2017. PMID: 28237978 Free PMC article.
-
Inhibition and avoidance of mRNA degradation by RNA viruses.Curr Opin Microbiol. 2012 Aug;15(4):500-5. doi: 10.1016/j.mib.2012.04.009. Epub 2012 May 23. Curr Opin Microbiol. 2012. PMID: 22626865 Free PMC article. Review.
-
The SARS-CoV-2 RNA-protein interactome in infected human cells.Nat Microbiol. 2021 Mar;6(3):339-353. doi: 10.1038/s41564-020-00846-z. Epub 2020 Dec 21. Nat Microbiol. 2021. PMID: 33349665 Free PMC article.
-
PCBP2 promotes the development of glioma by regulating FHL3/TGF-β/Smad signaling pathway.J Cell Physiol. 2020 Apr;235(4):3280-3291. doi: 10.1002/jcp.29104. Epub 2019 Nov 6. J Cell Physiol. 2020. PMID: 31693182 Free PMC article.
References
-
- Bartenschlager R, Sparacio S. Hepatitis C virus molecular clones and their replication capacity in vivo and in cell culture. Virus Res. 2007;127:195–207. - PubMed
-
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. - PubMed
-
- Di Bisceglie AM, Hoofnagle JH. Optimal therapy of hepatitis C. Hepatology. 2002;36:S121–S127. - PubMed
-
- Lindsay KL, Trepo C, Heintges T, Shiffman ML, Gordon SC, et al. A randomized, double-blind trial comparing PEGylated interferon alfa-2b to interferon alfa-2b as initial treatment for chronic hepatitis C. Hepatology. 2001;34:395–403. - PubMed
-
- Jordan JF, Jay HH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

