The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress
- PMID: 22021418
- PMCID: PMC3327187
- DOI: 10.1104/pp.111.188300
The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress
Abstract
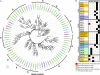
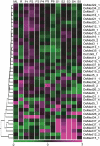
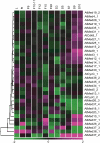
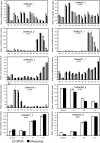
The Mediator (Med) complex relays regulatory information from DNA-bound transcription factors to the RNA polymerase II in eukaryotes. This macromolecular unit is composed of three core subcomplexes in addition to a separable kinase module. In this study, conservation of Meds has been investigated in 16 plant species representing seven diverse groups across the plant kingdom. Using Hidden Markov Model-based conserved motif searches, we have identified all the known yeast/metazoan Med components in one or more plant groups, including the Med26 subunits, which have not been reported so far for any plant species. We also detected orthologs for the Arabidopsis (Arabidopsis thaliana) Med32, -33, -34, -35, -36, and -37 in all the plant groups, and in silico analysis identified the Med32 and Med33 subunits as apparent orthologs of yeast/metazoan Med2/29 and Med5/24, respectively. Consequently, the plant Med complex appears to be composed of one or more members of 34 subunits, as opposed to 25 and 30 members in yeast and metazoans, respectively. Despite low similarity in primary Med sequences between the plants and their fungal/metazoan partners, secondary structure modeling of these proteins revealed a remarkable similarity between them, supporting the conservation of Med organization across kingdoms. Phylogenetic analysis between plant, human, and yeast revealed single clade relatedness for 29 Med genes families in plants, plant Meds being closer to human than to yeast counterparts. Expression profiling of rice (Oryza sativa) and Arabidopsis Med genes reveals that Meds not only act as a basal regulator of gene expression but may also have specific roles in plant development and under abiotic stress conditions.
Figures




Similar articles
-
Rice phospholipase A superfamily: organization, phylogenetic and expression analysis during abiotic stresses and development.PLoS One. 2012;7(2):e30947. doi: 10.1371/journal.pone.0030947. Epub 2012 Feb 17. PLoS One. 2012. PMID: 22363522 Free PMC article.
-
Spt-Ada-Gcn5-Acetyltransferase (SAGA) Complex in Plants: Genome Wide Identification, Evolutionary Conservation and Functional Determination.PLoS One. 2015 Aug 11;10(8):e0134709. doi: 10.1371/journal.pone.0134709. eCollection 2015. PLoS One. 2015. PMID: 26263547 Free PMC article.
-
MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress.BMC Genomics. 2007 Jul 18;8:242. doi: 10.1186/1471-2164-8-242. BMC Genomics. 2007. PMID: 17640358 Free PMC article.
-
Conservation and Divergence of Mediator Structure and Function: Insights from Plants.Plant Cell Physiol. 2017 Jan 1;58(1):4-21. doi: 10.1093/pcp/pcw176. Plant Cell Physiol. 2017. PMID: 28173572 Review.
-
Evolutionary expansion, functional diversification, and transcript profiling of plant Glutathione Peroxidases.Plant Sci. 2024 Apr;341:111991. doi: 10.1016/j.plantsci.2024.111991. Epub 2024 Jan 22. Plant Sci. 2024. PMID: 38266716 Review.
Cited by
-
Interaction between plastid and mitochondrial retrograde signalling pathways during changes to plastid redox status.Philos Trans R Soc Lond B Biol Sci. 2014 Mar 3;369(1640):20130231. doi: 10.1098/rstb.2013.0231. Print 2014 Apr 19. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 24591717 Free PMC article.
-
The characterization of Mediator 12 and 13 as conditional positive gene regulators in Arabidopsis.Nat Commun. 2020 Jun 3;11(1):2798. doi: 10.1038/s41467-020-16651-5. Nat Commun. 2020. PMID: 32493925 Free PMC article.
-
Comparative phylogenetic analysis of the mediator complex subunit in asparagus bean (Vigna unguiculata ssp. sesquipedialis) and its expression profile under cold stress.BMC Genomics. 2024 Feb 6;25(1):149. doi: 10.1186/s12864-024-10060-4. BMC Genomics. 2024. PMID: 38321384 Free PMC article.
-
Expression of AtMed15 of Arabidopsis in yeast causes flocculation and increases ethanol production in yeast culture.Sci Rep. 2016 Jun 16;6:27967. doi: 10.1038/srep27967. Sci Rep. 2016. PMID: 27306498 Free PMC article.
-
LACK OF SYMBIONT ACCOMMODATION controls intracellular symbiont accommodation in root nodule and arbuscular mycorrhizal symbiosis in Lotus japonicus.PLoS Genet. 2019 Jan 3;15(1):e1007865. doi: 10.1371/journal.pgen.1007865. eCollection 2019 Jan. PLoS Genet. 2019. PMID: 30605473 Free PMC article.
References
-
- Agarwal P, Arora R, Ray S, Singh AK, Singh VP, Takatsuji H, Kapoor S, Tyagi AK. (2007) Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol Biol 65: 467–485 - PubMed
-
- Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. (1999) Conserved structures of Mediator and RNA polymerase II holoenzyme. Science 283: 985–987 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

