Comprehensive analysis of microRNA genomic loci identifies pervasive repetitive-element origins
- PMID: 22016841
- PMCID: PMC3190270
- DOI: 10.4161/mge.1.1.15766
Comprehensive analysis of microRNA genomic loci identifies pervasive repetitive-element origins
Abstract
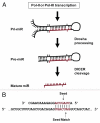
MicroRNAs (miRs) are small non-coding RNAs that generally function as negative regulators of target messenger RNAs (mRNAs) at the posttranscriptional level. MiRs bind to the 3'UTR of target mRNAs through complementary base pairing, resulting in target mRNA cleavage or translation repression. To date, over 15,000 distinct miRs have been identified in organisms ranging from viruses to man and interest in miR research continues to intensify. Of note, the most enlightening aspect of miR function-the mRNAs they target-continues to be elusive. Descriptions of the molecular origins of independent miR molecules currently support the hypothesis that miR hairpin generation is based on the adjacent insertion of two related transposable elements (TEs) at one genomic locus. Thus transcription across such TE interfaces establishes many, if not the majority of functional miRs. The implications of these findings are substantial for understanding how TEs confer increased genomic fitness, describing miR transcriptional regulations and making accurate miR target predictions. In this work, we have performed a comprehensive analysis of the genomic events responsible for the formation of all currently annotated miR loci. We find that the connection between miRs and transposable elements is more significant than previously appreciated, and more broadly, supports an important role for repetitive elements in miR origin, expression and regulatory network formation. Further, we demonstrate the utility of these findings in miR target prediction. Our results greatly expand the existing repertoire of defined miR origins, detailing the formation of 2,392 of 15,176 currently recognized miR genomic loci and supporting a mobile genetic element model for the genomic establishment of functional miRs.
Figures


Similar articles
-
OrbId: Origin-based identification of microRNA targets.Mob Genet Elements. 2012 Jul 1;2(4):184-192. doi: 10.4161/mge.21617. Mob Genet Elements. 2012. PMID: 23087843 Free PMC article.
-
Continuing analysis of microRNA origins: Formation from transposable element insertions and noncoding RNA mutations.Mob Genet Elements. 2013 Nov 1;3(6):e27755. doi: 10.4161/mge.27755. Epub 2014 Jan 10. Mob Genet Elements. 2013. PMID: 24475369 Free PMC article.
-
Domestication of transposable elements into MicroRNA genes in plants.PLoS One. 2011 May 3;6(5):e19212. doi: 10.1371/journal.pone.0019212. PLoS One. 2011. PMID: 21559273 Free PMC article.
-
MicroRNA and cancer--focus on apoptosis.J Cell Mol Med. 2009 Jan;13(1):12-23. doi: 10.1111/j.1582-4934.2008.00510.x. J Cell Mol Med. 2009. PMID: 19175697 Free PMC article. Review.
-
MicroRNAs in the immune response.Cytokine. 2008 Sep;43(3):391-4. doi: 10.1016/j.cyto.2008.07.016. Epub 2008 Aug 12. Cytokine. 2008. PMID: 18701320 Free PMC article. Review.
Cited by
-
OrbId: Origin-based identification of microRNA targets.Mob Genet Elements. 2012 Jul 1;2(4):184-192. doi: 10.4161/mge.21617. Mob Genet Elements. 2012. PMID: 23087843 Free PMC article.
-
Control by a hair's breadth: the role of microRNAs in the skin.Cell Mol Life Sci. 2013 Apr;70(7):1149-69. doi: 10.1007/s00018-012-1117-z. Epub 2012 Sep 15. Cell Mol Life Sci. 2013. PMID: 22983383 Free PMC article. Review.
-
Base Composition Characteristics of Mammalian miRNAs.J Nucleic Acids. 2013;2013:951570. doi: 10.1155/2013/951570. Epub 2013 Feb 24. J Nucleic Acids. 2013. PMID: 23710337 Free PMC article.
-
Functional microRNAs and target sites are created by lineage-specific transposition.Hum Mol Genet. 2014 Apr 1;23(7):1783-93. doi: 10.1093/hmg/ddt569. Epub 2013 Nov 13. Hum Mol Genet. 2014. PMID: 24234653 Free PMC article.
-
Role of microRNAs in lung development and pulmonary diseases.Pulm Circ. 2013 Apr;3(2):315-28. doi: 10.4103/2045-8932.114758. Pulm Circ. 2013. PMID: 24015331 Free PMC article. Review.
References
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
-
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. - PubMed
-
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
