RECQL5 cooperates with Topoisomerase II alpha in DNA decatenation and cell cycle progression
- PMID: 22013166
- PMCID: PMC3287182
- DOI: 10.1093/nar/gkr844
RECQL5 cooperates with Topoisomerase II alpha in DNA decatenation and cell cycle progression
Abstract
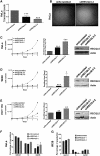
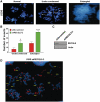
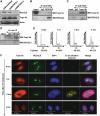
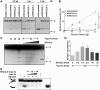
DNA decatenation mediated by Topoisomerase II is required to separate the interlinked sister chromatids post-replication. SGS1, a yeast homolog of the human RecQ family of helicases interacts with Topoisomerase II and plays a role in chromosome segregation, but this functional interaction has yet to be identified in higher organisms. Here, we report a physical and functional interaction of Topoisomerase IIα with RECQL5, one of five mammalian RecQ helicases, during DNA replication. Direct interaction of RECQL5 with Topoisomerase IIα stimulates the decatenation activity of Topoisomerase IIα. Consistent with these observations, RECQL5 co-localizes with Topoisomerase IIα during S-phase of the cell cycle. Moreover, cells with stable depletions of RECQL5 display a slow proliferation rate, a G2/M cell cycle arrest and late S-phase cycling defects. Metaphase spreads generated from RECQL5-depleted cells exhibit undercondensed and entangled chromosomes. Further, RECQL5-depleted cells activate a G2/M checkpoint and undergo apoptosis. These phenotypes are similar to those observed when Topoisomerase II catalytic activity is inhibited. These results reveal an important role for RECQL5 in the maintenance of genomic stability and a new insight into the decatenation process.
Figures



Similar articles
-
BAF complexes facilitate decatenation of DNA by topoisomerase IIα.Nature. 2013 May 30;497(7451):624-7. doi: 10.1038/nature12146. Epub 2013 May 22. Nature. 2013. PMID: 23698369 Free PMC article.
-
Bloom's syndrome and PICH helicases cooperate with topoisomerase IIα in centromere disjunction before anaphase.PLoS One. 2012;7(4):e33905. doi: 10.1371/journal.pone.0033905. Epub 2012 Apr 26. PLoS One. 2012. PMID: 22563370 Free PMC article.
-
The SET and transposase domain protein Metnase enhances chromosome decatenation: regulation by automethylation.Nucleic Acids Res. 2008 Oct;36(18):5822-31. doi: 10.1093/nar/gkn560. Epub 2008 Sep 12. Nucleic Acids Res. 2008. PMID: 18790802 Free PMC article.
-
Human RECQL5: guarding the crossroads of DNA replication and transcription and providing backup capability.Crit Rev Biochem Mol Biol. 2013 May-Jun;48(3):289-99. doi: 10.3109/10409238.2013.792770. Epub 2013 Apr 29. Crit Rev Biochem Mol Biol. 2013. PMID: 23627586 Free PMC article. Review.
-
Sister chromatid decatenation: bridging the gaps in our knowledge.Cell Cycle. 2015;14(19):3040-4. doi: 10.1080/15384101.2015.1078039. Cell Cycle. 2015. PMID: 26266709 Free PMC article. Review.
Cited by
-
RECQL5 at the Intersection of Replication and Transcription.Front Cell Dev Biol. 2020 May 25;8:324. doi: 10.3389/fcell.2020.00324. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32523948 Free PMC article. Review.
-
The RECG1 DNA Translocase Is a Key Factor in Recombination Surveillance, Repair, and Segregation of the Mitochondrial DNA in Arabidopsis.Plant Cell. 2015 Oct;27(10):2907-25. doi: 10.1105/tpc.15.00680. Epub 2015 Oct 13. Plant Cell. 2015. PMID: 26462909 Free PMC article.
-
BAF complexes facilitate decatenation of DNA by topoisomerase IIα.Nature. 2013 May 30;497(7451):624-7. doi: 10.1038/nature12146. Epub 2013 May 22. Nature. 2013. PMID: 23698369 Free PMC article.
-
Clinically Applicable Inhibitors Impacting Genome Stability.Molecules. 2018 May 13;23(5):1166. doi: 10.3390/molecules23051166. Molecules. 2018. PMID: 29757235 Free PMC article. Review.
-
Recruitment and retention dynamics of RECQL5 at DNA double strand break sites.DNA Repair (Amst). 2012 Jul 1;11(7):624-35. doi: 10.1016/j.dnarep.2012.05.001. Epub 2012 May 24. DNA Repair (Amst). 2012. PMID: 22633600 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

