Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma
- PMID: 22006338
- PMCID: PMC3203814
- DOI: 10.1073/pnas.1114669108
Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma
Abstract
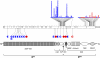
Squamous cell carcinomas (SCCs) are one of the most frequent forms of human malignancy, but, other than TP53 mutations, few causative somatic aberrations have been identified. We identified NOTCH1 or NOTCH2 mutations in ~75% of cutaneous SCCs and in a lesser fraction of lung SCCs, defining a spectrum for the most prevalent tumor suppressor specific to these epithelial malignancies. Notch receptors normally transduce signals in response to ligands on neighboring cells, regulating metazoan lineage selection and developmental patterning. Our findings therefore illustrate a central role for disruption of microenvironmental communication in cancer progression. NOTCH aberrations include frameshift and nonsense mutations leading to receptor truncations as well as point substitutions in key functional domains that abrogate signaling in cell-based assays. Oncogenic gain-of-function mutations in NOTCH1 commonly occur in human T-cell lymphoblastic leukemia/lymphoma and B-cell chronic lymphocytic leukemia. The bifunctional role of Notch in human cancer thus emphasizes the context dependency of signaling outcomes and suggests that targeted inhibition of the Notch pathway may induce squamous epithelial malignancies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

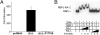
Similar articles
-
Notch Signaling Components: Diverging Prognostic Indicators in Lung Adenocarcinoma.Medicine (Baltimore). 2016 May;95(20):e3715. doi: 10.1097/MD.0000000000003715. Medicine (Baltimore). 2016. PMID: 27196489 Free PMC article.
-
Genomic analysis of metastatic cutaneous squamous cell carcinoma.Clin Cancer Res. 2015 Mar 15;21(6):1447-56. doi: 10.1158/1078-0432.CCR-14-1773. Epub 2015 Jan 14. Clin Cancer Res. 2015. PMID: 25589618 Free PMC article.
-
Mutational analysis of NOTCH1, 2, 3 and 4 genes in common solid cancers and acute leukemias.APMIS. 2007 Dec;115(12):1357-63. doi: 10.1111/j.1600-0463.2007.00751.x. APMIS. 2007. PMID: 18184405
-
Does Notch play a tumor suppressor role across diverse squamous cell carcinomas?Cancer Med. 2016 Aug;5(8):2048-60. doi: 10.1002/cam4.731. Epub 2016 May 26. Cancer Med. 2016. PMID: 27228302 Free PMC article. Review.
-
The NOTCH Pathway in Head and Neck Squamous Cell Carcinoma.J Dent Res. 2018 Jun;97(6):645-653. doi: 10.1177/0022034518760297. Epub 2018 Feb 28. J Dent Res. 2018. PMID: 29489439 Free PMC article. Review.
Cited by
-
BMP-binding protein twisted gastrulation is required in mammary gland epithelium for normal ductal elongation and myoepithelial compartmentalization.Dev Biol. 2013 Jan 1;373(1):95-106. doi: 10.1016/j.ydbio.2012.10.007. Epub 2012 Oct 24. Dev Biol. 2013. PMID: 23103586 Free PMC article.
-
Identification of potential gene drivers of cutaneous squamous cell carcinoma: Analysis of microarray data.Medicine (Baltimore). 2020 Sep 25;99(39):e22257. doi: 10.1097/MD.0000000000022257. Medicine (Baltimore). 2020. PMID: 32991423 Free PMC article.
-
Therapeutic Approaches for Non-Melanoma Skin Cancer: Standard of Care and Emerging Modalities.Int J Mol Sci. 2024 Jun 27;25(13):7056. doi: 10.3390/ijms25137056. Int J Mol Sci. 2024. PMID: 39000164 Free PMC article. Review.
-
Oncogenic and Tumor-Suppressive Functions of NOTCH Signaling in Glioma.Cells. 2020 Oct 15;9(10):2304. doi: 10.3390/cells9102304. Cells. 2020. PMID: 33076453 Free PMC article. Review.
-
Mutations in POFUT1, encoding protein O-fucosyltransferase 1, cause generalized Dowling-Degos disease.Am J Hum Genet. 2013 Jun 6;92(6):895-903. doi: 10.1016/j.ajhg.2013.04.022. Epub 2013 May 16. Am J Hum Genet. 2013. PMID: 23684010 Free PMC article.
References
-
- Madan V, Lear JT, Szeimies R-M. Non-melanoma skin cancer. Lancet. 2010;375:673–685. - PubMed
-
- Janes SM, Watt FM. New roles for integrins in squamous-cell carcinoma. Nat Rev Cancer. 2006;6:175–183. - PubMed
-
- Shigematsu H, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. - PubMed
-
- Kemp CJ, Donehower LA, Bradley A, Balmain A. Reduction of p53 gene dosage does not increase initiation or promotion but enhances malignant progression of chemically induced skin tumors. Cell. 1993;74:813–822. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08 CA169865/CA/NCI NIH HHS/United States
- R01 AR051930/AR/NIAMS NIH HHS/United States
- K08 CA137153/CA/NCI NIH HHS/United States
- U24 CA126551/CA/NCI NIH HHS/United States
- R01 CA092433/CA/NCI NIH HHS/United States
- U24 CA143799/CA/NCI NIH HHS/United States
- U54 CA 112970/CA/NCI NIH HHS/United States
- 13044/CRUK_/Cancer Research UK/United Kingdom
- P50 CA 58207/CA/NCI NIH HHS/United States
- P50 CA058207/CA/NCI NIH HHS/United States
- KO8CA137153/CA/NCI NIH HHS/United States
- U54 CA112970/CA/NCI NIH HHS/United States
- P50 CA121974/CA/NCI NIH HHS/United States
- 5KO8AR057763/AR/NIAMS NIH HHS/United States
- AR051930/AR/NIAMS NIH HHS/United States
- P01 CA119070/CA/NCI NIH HHS/United States
- U24 CA1437991/CA/NCI NIH HHS/United States
- K08 AR057763/AR/NIAMS NIH HHS/United States
- R01AG028492/AG/NIA NIH HHS/United States
- R01 AG028492/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

