The curious chemical biology of cytosine: deamination, methylation, and oxidation as modulators of genomic potential
- PMID: 22004246
- PMCID: PMC3262930
- DOI: 10.1021/cb2002895
The curious chemical biology of cytosine: deamination, methylation, and oxidation as modulators of genomic potential
Abstract
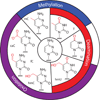
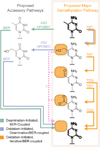
A multitude of functions have evolved around cytosine within DNA, endowing the base with physiological significance beyond simple information storage. This versatility arises from enzymes that chemically modify cytosine to expand the potential of the genome. Some modifications alter coding sequences, such as deamination of cytosine by AID/APOBEC enzymes to generate immunologic or virologic diversity. Other modifications are critical to epigenetic control, altering gene expression or cellular identity. Of these, cytosine methylation is well understood, in contrast to recently discovered modifications, such as oxidation by TET enzymes to 5-hydroxymethylcytosine. Further complexity results from cytosine demethylation, an enigmatic process that impacts cellular pluripotency. Recent insights help us to propose an integrated DNA demethylation model, accounting for contributions from cytosine oxidation, deamination, and base excision repair. Taken together, this rich medley of alterations renders cytosine a genomic "wild card", whose context-dependent functions make the base far more than a static letter in the code of life.
Figures
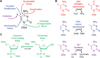

Similar articles
-
Expanding the epigenetic landscape: novel modifications of cytosine in genomic DNA.Cold Spring Harb Perspect Biol. 2014 Oct 1;6(10):a018630. doi: 10.1101/cshperspect.a018630. Cold Spring Harb Perspect Biol. 2014. PMID: 25274704 Free PMC article. Review.
-
[Oxidation and deamination of nucleobases as an epigenetic tool].Postepy Hig Med Dosw (Online). 2012 May 24;66:275-86. doi: 10.5604/17322693.997954. Postepy Hig Med Dosw (Online). 2012. PMID: 22706113 Review. Polish.
-
TET enzymatic oxidation of 5-methylcytosine, 5-hydroxymethylcytosine and 5-formylcytosine.Mutat Res Genet Toxicol Environ Mutagen. 2014 Apr;764-765:18-35. doi: 10.1016/j.mrgentox.2013.09.001. Epub 2013 Sep 14. Mutat Res Genet Toxicol Environ Mutagen. 2014. PMID: 24045206 Review.
-
Epigenetic modifications in DNA could mimic oxidative DNA damage: A double-edged sword.DNA Repair (Amst). 2015 Aug;32:52-57. doi: 10.1016/j.dnarep.2015.04.013. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956859 Review.
-
Oxidized C5-methyl cytosine bases in DNA: 5-Hydroxymethylcytosine; 5-formylcytosine; and 5-carboxycytosine.Free Radic Biol Med. 2017 Jun;107:62-68. doi: 10.1016/j.freeradbiomed.2016.11.038. Epub 2016 Nov 24. Free Radic Biol Med. 2017. PMID: 27890639 Review.
Cited by
-
High UV damage and low repair, but not cytosine deamination, stimulate mutation hotspots at ETS binding sites in melanoma.Proc Natl Acad Sci U S A. 2024 Jan 23;121(4):e2310854121. doi: 10.1073/pnas.2310854121. Epub 2024 Jan 19. Proc Natl Acad Sci U S A. 2024. PMID: 38241433 Free PMC article.
-
Is DNA methylation in the brain a mechanism of alcohol use disorder?Front Behav Neurosci. 2023 Jan 26;17:957203. doi: 10.3389/fnbeh.2023.957203. eCollection 2023. Front Behav Neurosci. 2023. PMID: 36778133 Free PMC article. Review.
-
A fluorescent reporter for quantification and enrichment of DNA editing by APOBEC-Cas9 or cleavage by Cas9 in living cells.Nucleic Acids Res. 2018 Aug 21;46(14):e84. doi: 10.1093/nar/gky332. Nucleic Acids Res. 2018. PMID: 29746667 Free PMC article.
-
Mechanisms for enzymatic cleavage of the N-glycosidic bond in DNA.Org Biomol Chem. 2014 Nov 14;12(42):8367-78. doi: 10.1039/c4ob01063a. Org Biomol Chem. 2014. PMID: 25181003 Free PMC article. Review.
-
Reversible DNA-Protein Cross-Linking at Epigenetic DNA Marks.Angew Chem Int Ed Engl. 2017 Nov 6;56(45):14130-14134. doi: 10.1002/anie.201708286. Epub 2017 Oct 6. Angew Chem Int Ed Engl. 2017. PMID: 28898504 Free PMC article.
References
-
- Kohli RM. Grand Challenge Commentary: The Chemistry of a Dynamic Genome. Nat. Chem. Biol. 2010;6:866–868. - PubMed
-
- Grosjean H. Nucleic Acids Are Not Boring Long Polymers of Only Four Types of Nucleotides: A Guided Tour. In: Grosjean H, editor. DNA and RNA Modification Enzymes. Austin: Landes Bioscience; 2009. pp. 1–18.
-
- Gerber AP, Keller W. RNA Editing by Base Deamination: More Enzymes, More Targets, New Mysteries. Trends Biochem. Sci. 2001;26:376–384. - PubMed
-
- Gott JM, Emeson RB. Functions and Mechanisms of RNA Editing. Annu. Rev. Genet. 2000;34:499–531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

