An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma
- PMID: 22000015
- PMCID: PMC3214599
- DOI: 10.1016/j.cell.2011.09.041
An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma
Abstract
By analyzing gene expression data in glioblastoma in combination with matched microRNA profiles, we have uncovered a posttranscriptional regulation layer of surprising magnitude, comprising more than 248,000 microRNA (miR)-mediated interactions. These include ∼7,000 genes whose transcripts act as miR "sponges" and 148 genes that act through alternative, nonsponge interactions. Biochemical analyses in cell lines confirmed that this network regulates established drivers of tumor initiation and subtype implementation, including PTEN, PDGFRA, RB1, VEGFA, STAT3, and RUNX1, suggesting that these interactions mediate crosstalk between canonical oncogenic pathways. siRNA silencing of 13 miR-mediated PTEN regulators, whose locus deletions are predictive of PTEN expression variability, was sufficient to downregulate PTEN in a 3'UTR-dependent manner and to increase tumor cell growth rates. Thus, miR-mediated interactions provide a mechanistic, experimentally validated rationale for the loss of PTEN expression in a large number of glioma samples with an intact PTEN locus.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
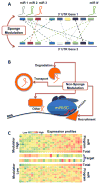

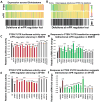
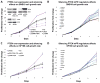
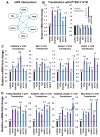

Comment in
-
Epigenetics. Layer by layer.Nat Rev Cancer. 2011 Nov 3;11(12):830. doi: 10.1038/nrc3172. Nat Rev Cancer. 2011. PMID: 22048565 No abstract available.
-
Regulatory RNA: layer by layer.Nat Rev Genet. 2011 Nov 3;12(12):804. doi: 10.1038/nrg3108. Nat Rev Genet. 2011. PMID: 22048663 No abstract available.
-
RNA: a new layer of regulation.Nat Rev Mol Cell Biol. 2011 Nov 3;12(12):766. doi: 10.1038/nrm3225. Nat Rev Mol Cell Biol. 2011. PMID: 22048709 No abstract available.
-
ceRNAs: miRNA target mimic mimics.Cell. 2011 Dec 23;147(7):1431-2. doi: 10.1016/j.cell.2011.12.003. Cell. 2011. PMID: 22196719 No abstract available.
Similar articles
-
PDGF-B-mediated downregulation of miR-21: new insights into PDGF signaling in glioblastoma.Hum Mol Genet. 2012 Dec 1;21(23):5118-30. doi: 10.1093/hmg/dds358. Epub 2012 Aug 24. Hum Mol Genet. 2012. PMID: 22922228 Free PMC article.
-
Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship.Proc Natl Acad Sci U S A. 2010 Feb 2;107(5):2183-8. doi: 10.1073/pnas.0909896107. Epub 2010 Jan 13. Proc Natl Acad Sci U S A. 2010. PMID: 20080666 Free PMC article.
-
MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol.BMC Cancer. 2010 Jan 31;10:27. doi: 10.1186/1471-2407-10-27. BMC Cancer. 2010. PMID: 20113523 Free PMC article.
-
Dickkopf-1 (DKK1) phosphatase and tensin homolog on chromosome 10 (PTEN) crosstalk via microRNA interference in the diabetic heart.Basic Res Cardiol. 2013 May;108(3):352. doi: 10.1007/s00395-013-0352-2. Epub 2013 May 1. Basic Res Cardiol. 2013. PMID: 23636253
-
MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN.BMC Cancer. 2010 Jul 12;10:367. doi: 10.1186/1471-2407-10-367. BMC Cancer. 2010. PMID: 20618998 Free PMC article.
Cited by
-
Systematic exploration of autonomous modules in noisy microRNA-target networks for testing the generality of the ceRNA hypothesis.BMC Genomics. 2014 Dec 24;15(1):1178. doi: 10.1186/1471-2164-15-1178. BMC Genomics. 2014. PMID: 25539629 Free PMC article.
-
Subpathway-LNCE: Identify dysfunctional subpathways competitively regulated by lncRNAs through integrating lncRNA-mRNA expression profile and pathway topologies.Oncotarget. 2016 Oct 25;7(43):69857-69870. doi: 10.18632/oncotarget.12005. Oncotarget. 2016. PMID: 27634882 Free PMC article.
-
A frequent somatic mutation in the 3'UTR of GAPDH facilitates the development of ovarian cancer by creating a miR‑125b binding site.Oncol Rep. 2020 Sep;44(3):887-896. doi: 10.3892/or.2020.7663. Epub 2020 Jun 25. Oncol Rep. 2020. PMID: 32705257 Free PMC article.
-
Competing Endogenous RNA Networks in Glioma.Front Genet. 2021 Apr 29;12:675498. doi: 10.3389/fgene.2021.675498. eCollection 2021. Front Genet. 2021. PMID: 33995499 Free PMC article. Review.
-
Complexity and Specificity of the Neutrophil Transcriptomes in Juvenile Idiopathic Arthritis.Sci Rep. 2016 Jun 7;6:27453. doi: 10.1038/srep27453. Sci Rep. 2016. PMID: 27271962 Free PMC article.
References
-
- Barrat A, Barthelemy M, Vespignani A. Dynamical processes on complex networks. Cambridge, UK, New York: Cambridge University Press; 2008.
-
- Cesana, et al. this issue of Cell. 2011.
-
- Chatterjee S, Fasler M, Bussing I, Grosshans H. Target-mediated protection of endogenous microRNAs in C. elegans. Dev Cell. 2011;20:388–396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

