The schizophrenia susceptibility factor dysbindin and its associated complex sort cargoes from cell bodies to the synapse
- PMID: 21998198
- PMCID: PMC3237628
- DOI: 10.1091/mbc.E11-07-0592
The schizophrenia susceptibility factor dysbindin and its associated complex sort cargoes from cell bodies to the synapse
Abstract
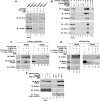
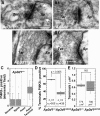
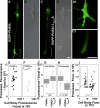
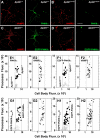
Dysbindin assembles into the biogenesis of lysosome-related organelles complex 1 (BLOC-1), which interacts with the adaptor protein complex 3 (AP-3), mediating a common endosome-trafficking route. Deficiencies in AP-3 and BLOC-1 affect synaptic vesicle composition. However, whether AP-3-BLOC-1-dependent sorting events that control synapse membrane protein content take place in cell bodies upstream of nerve terminals remains unknown. We tested this hypothesis by analyzing the targeting of phosphatidylinositol-4-kinase type II α (PI4KIIα), a membrane protein present in presynaptic and postsynaptic compartments. PI4KIIα copurified with BLOC-1 and AP-3 in neuronal cells. These interactions translated into a decreased PI4KIIα content in the dentate gyrus of dysbindin-null BLOC-1 deficiency and AP-3-null mice. Reduction of PI4KIIα in the dentate reflects a failure to traffic from the cell body. PI4KIIα was targeted to processes in wild-type primary cultured cortical neurons and PC12 cells but failed to reach neurites in cells lacking either AP-3 or BLOC-1. Similarly, disruption of an AP-3-sorting motif in PI4KIIα impaired its sorting into processes of PC12 and primary cultured cortical neuronal cells. Our findings indicate a novel vesicle transport mechanism requiring BLOC-1 and AP-3 complexes for cargo sorting from neuronal cell bodies to neurites and nerve terminals.
Figures




Similar articles
-
Dysbindin-containing complexes and their proposed functions in brain: from zero to (too) many in a decade.ASN Neuro. 2011 May 27;3(2):e00058. doi: 10.1042/AN20110010. ASN Neuro. 2011. PMID: 21504412 Free PMC article. Review.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
MeCP2 regulates the synaptic expression of a Dysbindin-BLOC-1 network component in mouse brain and human induced pluripotent stem cell-derived neurons.PLoS One. 2013 Jun 4;8(6):e65069. doi: 10.1371/journal.pone.0065069. Print 2013. PLoS One. 2013. PMID: 23750231 Free PMC article.
-
Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. Update in: Commun Chem. 2024 Nov 19;7(1):271. doi: 10.1038/s42004-024-01352-7. PMID: 38260445 Free PMC article. Updated. Preprint.
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
Cited by
-
Type II phosphatidylinositol 4-kinases function sequentially in cargo delivery from early endosomes to melanosomes.J Cell Biol. 2022 Nov 7;221(11):e202110114. doi: 10.1083/jcb.202110114. Epub 2022 Sep 28. J Cell Biol. 2022. PMID: 36169639 Free PMC article.
-
Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder.Mol Psychiatry. 2015 May;20(5):563-572. doi: 10.1038/mp.2014.82. Epub 2014 Aug 12. Mol Psychiatry. 2015. PMID: 25113377 Free PMC article.
-
The Endosome Localized Arf-GAP AGAP1 Modulates Dendritic Spine Morphology Downstream of the Neurodevelopmental Disorder Factor Dysbindin.Front Cell Neurosci. 2016 Sep 22;10:218. doi: 10.3389/fncel.2016.00218. eCollection 2016. Front Cell Neurosci. 2016. PMID: 27713690 Free PMC article.
-
Phosphatidylinositol 4-kinases and PI4P metabolism in the nervous system: roles in psychiatric and neurological diseases.Mol Neurobiol. 2013 Feb;47(1):361-72. doi: 10.1007/s12035-012-8358-6. Epub 2012 Oct 10. Mol Neurobiol. 2013. PMID: 23054682 Review.
-
Coupling of microtubule motors with AP-3 generated organelles in axons by NEEP21 family member calcyon.Mol Biol Cell. 2018 Aug 15;29(17):2055-2068. doi: 10.1091/mbc.E18-01-0007. Epub 2018 Jun 27. Mol Biol Cell. 2018. PMID: 29949458 Free PMC article.
References
-
- Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. - PubMed
-
- Bhardwaj SK, Baharnoori M, Sharif-Askari B, Kamath A, Williams S, Srivastava LK. Behavioral characterization of dysbindin-1 deficient sandy mice. Behav Brain Res. 2009;197:435–441. - PubMed
-
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. - PubMed
-
- Borck G, et al. Clinical, cellular, and neuropathological consequences of AP1S2 mutations: further delineation of a recognizable X-linked mental retardation syndrome. Hum Mutat. 2008;29:966–974. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

