Analysis of subcellular G3BP redistribution during rubella virus infection
- PMID: 21994324
- PMCID: PMC3352340
- DOI: 10.1099/vir.0.036780-0
Analysis of subcellular G3BP redistribution during rubella virus infection
Abstract
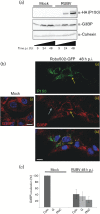
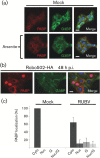
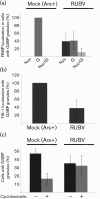
Rubella virus (RUBV) replicates slowly and to low titre in vertebrate cultured cells, with minimal cytopathology. To determine whether a cellular stress response is induced during such an infection, the formation of Ras-GAP-SH3 domain-binding protein (G3BP)-containing stress granules (SGs) in RUBV-infected cells was examined. Late in infection, accumulation of G3BP granules was detected, albeit in fewer than half of infected cells. Active virus RNA replication was required for induction of these granules, but they were found to differ from SGs induced by arsenite treatment both in composition (they did not uniformly contain other SG proteins, such as PABP and TIA-1) and in resistance to cycloheximide treatment. Thus, bona fide SGs do not appear to be induced during RUBV infection. The distribution of G3BP, either on its own or in granules, did not overlap with that of dsRNA-containing replication complexes, indicating that it played no role in virus RNA synthesis. However, G3BP did co-localize with viral ssRNAs in perinuclear clusters, suggesting an interaction that could possibly be important in a post-replicative role in virus replication, such as encapsidation.
Figures
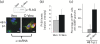

Similar articles
-
Herpes simplex virus 2 infection impacts stress granule accumulation.J Virol. 2012 Aug;86(15):8119-30. doi: 10.1128/JVI.00313-12. Epub 2012 May 23. J Virol. 2012. PMID: 22623775 Free PMC article.
-
Newcastle disease virus induces stable formation of bona fide stress granules to facilitate viral replication through manipulating host protein translation.FASEB J. 2017 Apr;31(4):1337-1353. doi: 10.1096/fj.201600980R. Epub 2016 Dec 23. FASEB J. 2017. PMID: 28011649
-
Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication.J Virol. 2015 Apr;89(8):4457-69. doi: 10.1128/JVI.03612-14. Epub 2015 Feb 4. J Virol. 2015. PMID: 25653451 Free PMC article.
-
Research Progress on the Structure and Function of G3BP.Front Immunol. 2021 Aug 30;12:718548. doi: 10.3389/fimmu.2021.718548. eCollection 2021. Front Immunol. 2021. PMID: 34526993 Free PMC article. Review.
-
Role(s) of G3BPs in Human Pathogenesis.J Pharmacol Exp Ther. 2023 Oct;387(1):100-110. doi: 10.1124/jpet.122.001538. Epub 2023 Jul 19. J Pharmacol Exp Ther. 2023. PMID: 37468286 Free PMC article. Review.
Cited by
-
HIV-2 genomic RNA accumulates in stress granules in the absence of active translation.Nucleic Acids Res. 2014 Nov 10;42(20):12861-75. doi: 10.1093/nar/gku1017. Epub 2014 Oct 28. Nucleic Acids Res. 2014. PMID: 25352557 Free PMC article.
-
Binding of cellular p32 protein to the rubella virus P150 replicase protein via PxxPxR motifs.J Gen Virol. 2012 Apr;93(Pt 4):807-816. doi: 10.1099/vir.0.038901-0. Epub 2012 Jan 13. J Gen Virol. 2012. PMID: 22238231 Free PMC article.
-
Who Regulates Whom? An Overview of RNA Granules and Viral Infections.Viruses. 2016 Jun 28;8(7):180. doi: 10.3390/v8070180. Viruses. 2016. PMID: 27367717 Free PMC article. Review.
-
Rasputin a decade on and more promiscuous than ever? A review of G3BPs.Biochim Biophys Acta Mol Cell Res. 2019 Mar;1866(3):360-370. doi: 10.1016/j.bbamcr.2018.09.001. Epub 2018 Sep 5. Biochim Biophys Acta Mol Cell Res. 2019. PMID: 30595162 Free PMC article. Review.
-
Stress granules: potential therapeutic targets for infectious and inflammatory diseases.Front Immunol. 2023 May 2;14:1145346. doi: 10.3389/fimmu.2023.1145346. eCollection 2023. Front Immunol. 2023. PMID: 37205103 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

