A SILAC-based screen for Methyl-CpG binding proteins identifies RBP-J as a DNA methylation and sequence-specific binding protein
- PMID: 21991380
- PMCID: PMC3185043
- DOI: 10.1371/journal.pone.0025884
A SILAC-based screen for Methyl-CpG binding proteins identifies RBP-J as a DNA methylation and sequence-specific binding protein
Abstract
Background: DNA methylation is an epigenetic modification that plays a crucial role in a variety of biological processes. Methylated DNA is specifically bound by Methyl-CpG Binding Proteins (MBPs). Three different types of MBPs have been identified so far: the Methyl-CpG Binding Domain (MBD) family proteins, three BTB/POZ-Zn-finger proteins, and UHRF1. Most of the known MBPs have been identified via homology with the MBD and Zn-finger domains as present in MeCP2 and Kaiso, respectively. It is conceivable that other proteins are capable of recognizing methylated DNA.
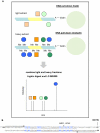
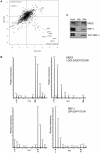
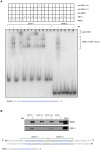
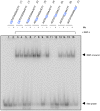
Methodology/principal findings: For the purpose of identifying novel 'readers' we set up a methyl-CpG pull-down assay combined with stable-isotope labeling by amino acids in cell culture (SILAC). In a methyl-CpG pull-down with U937 nuclear extracts, we recovered several known MBPs and almost all subunits of the MBD2/NuRD complex as methylation specific binders, providing proof-of-principle. Interestingly, RBP-J, the transcription factor downstream of Notch receptors, also bound the DNA in a methylation dependent manner. Follow-up pull-downs and electrophoretic mobility shift assays (EMSAs) showed that RBP-J binds methylated DNA in the context of a mutated RBP-J consensus motif.
Conclusions/significance: The here described SILAC/methyl-CpG pull-down constitutes a new approach to identify potential novel DNAme readers and will advance unraveling of the complete methyl-DNA interactome.
Conflict of interest statement
Figures

Similar articles
-
MBD3L2 interacts with MBD3 and components of the NuRD complex and can oppose MBD2-MeCP1-mediated methylation silencing.J Biol Chem. 2005 Apr 1;280(13):12700-9. doi: 10.1074/jbc.M413492200. Epub 2005 Jan 27. J Biol Chem. 2005. PMID: 15701600
-
On how mammalian transcription factors recognize methylated DNA.Epigenetics. 2013 Feb;8(2):131-7. doi: 10.4161/epi.23632. Epub 2013 Jan 16. Epigenetics. 2013. PMID: 23324617 Free PMC article. Review.
-
The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides.Nucleic Acids Res. 2002 Jul 1;30(13):2911-9. doi: 10.1093/nar/gkf398. Nucleic Acids Res. 2002. PMID: 12087177 Free PMC article.
-
Zinc Finger Readers of Methylated DNA.Molecules. 2018 Oct 7;23(10):2555. doi: 10.3390/molecules23102555. Molecules. 2018. PMID: 30301273 Free PMC article. Review.
-
Methyl-CpG-binding domain proteins: readers of the epigenome.Epigenomics. 2015;7(6):1051-73. doi: 10.2217/epi.15.39. Epub 2015 Apr 30. Epigenomics. 2015. PMID: 25927341 Review.
Cited by
-
Pleiotropic Role of Notch Signaling in Human Skin Diseases.Int J Mol Sci. 2020 Jun 13;21(12):4214. doi: 10.3390/ijms21124214. Int J Mol Sci. 2020. PMID: 32545758 Free PMC article. Review.
-
Making the most of methylation.Elife. 2013 Sep 3;2:e01387. doi: 10.7554/eLife.01387. Elife. 2013. PMID: 24015362 Free PMC article.
-
Epigenetic Therapeutics Targeting NRF2/KEAP1 Signaling in Cancer Oxidative Stress.Front Pharmacol. 2022 Jun 9;13:924817. doi: 10.3389/fphar.2022.924817. eCollection 2022. Front Pharmacol. 2022. PMID: 35754474 Free PMC article. Review.
-
A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation.Genome Biol. 2013;14(10):R119. doi: 10.1186/gb-2013-14-10-r119. Genome Biol. 2013. PMID: 24156278 Free PMC article.
-
DNA Methylation and Adult Neurogenesis.Brain Plast. 2017 Nov 9;3(1):5-26. doi: 10.3233/BPL-160034. Brain Plast. 2017. PMID: 29765857 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

