A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria
- PMID: 21987634
- PMCID: PMC3198156
- DOI: 10.1083/jcb.201107053
A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria
Abstract
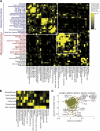
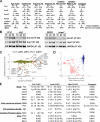
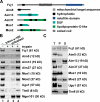
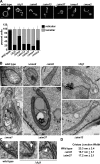
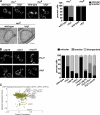
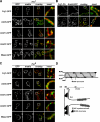
To broadly explore mitochondrial structure and function as well as the communication of mitochondria with other cellular pathways, we constructed a quantitative, high-density genetic interaction map (the MITO-MAP) in Saccharomyces cerevisiae. The MITO-MAP provides a comprehensive view of mitochondrial function including insights into the activity of uncharacterized mitochondrial proteins and the functional connection between mitochondria and the ER. The MITO-MAP also reveals a large inner membrane-associated complex, which we term MitOS for mitochondrial organizing structure, comprised of Fcj1/Mitofilin, a conserved inner membrane protein, and five additional components. MitOS physically and functionally interacts with both outer and inner membrane components and localizes to extended structures that wrap around the inner membrane. We show that MitOS acts in concert with ATP synthase dimers to organize the inner membrane and promote normal mitochondrial morphology. We propose that MitOS acts as a conserved mitochondrial skeletal structure that differentiates regions of the inner membrane to establish the normal internal architecture of mitochondria.
Figures
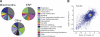
Similar articles
-
MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization.Mol Biol Cell. 2012 Jan;23(2):247-57. doi: 10.1091/mbc.E11-09-0774. Epub 2011 Nov 23. Mol Biol Cell. 2012. PMID: 22114354 Free PMC article.
-
The C-terminal domain of Fcj1 is required for formation of crista junctions and interacts with the TOB/SAM complex in mitochondria.Mol Biol Cell. 2012 Jun;23(11):2143-55. doi: 10.1091/mbc.E11-10-0831. Epub 2012 Apr 11. Mol Biol Cell. 2012. PMID: 22496419 Free PMC article.
-
Mitochondrial contact site and cristae organizing system.Curr Opin Cell Biol. 2016 Aug;41:33-42. doi: 10.1016/j.ceb.2016.03.013. Epub 2016 Apr 7. Curr Opin Cell Biol. 2016. PMID: 27062547 Review.
-
Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane.Mol Biol Cell. 2012 Oct;23(20):3948-56. doi: 10.1091/mbc.E12-04-0295. Epub 2012 Aug 23. Mol Biol Cell. 2012. PMID: 22918945 Free PMC article.
-
Mitofilin complexes: conserved organizers of mitochondrial membrane architecture.Biol Chem. 2012 Nov;393(11):1247-61. doi: 10.1515/hsz-2012-0239. Biol Chem. 2012. PMID: 23109542 Review.
Cited by
-
MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization.Mol Biol Cell. 2012 Jan;23(2):247-57. doi: 10.1091/mbc.E11-09-0774. Epub 2011 Nov 23. Mol Biol Cell. 2012. PMID: 22114354 Free PMC article.
-
Mitochondrial protein synthesis, import, and assembly.Genetics. 2012 Dec;192(4):1203-34. doi: 10.1534/genetics.112.141267. Genetics. 2012. PMID: 23212899 Free PMC article.
-
MICOS and phospholipid transfer by Ups2-Mdm35 organize membrane lipid synthesis in mitochondria.J Cell Biol. 2016 Jun 6;213(5):525-34. doi: 10.1083/jcb.201602007. Epub 2016 May 30. J Cell Biol. 2016. PMID: 27241913 Free PMC article.
-
MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation.EMBO J. 2020 Jul 15;39(14):e104105. doi: 10.15252/embj.2019104105. Epub 2020 Jun 22. EMBO J. 2020. PMID: 32567732 Free PMC article.
-
Disrupted in schizophrenia 1 (DISC1) is a constituent of the mammalian mitochondrial contact site and cristae organizing system (MICOS) complex, and is essential for oxidative phosphorylation.Hum Mol Genet. 2016 Oct 1;25(19):4157-4169. doi: 10.1093/hmg/ddw250. Epub 2016 Jul 27. Hum Mol Genet. 2016. PMID: 27466199 Free PMC article.
References
-
- Ackerman S.H., Tzagoloff A. 1990. ATP10, a yeast nuclear gene required for the assembly of the mitochondrial F1-F0 complex. J. Biol. Chem. 265:9952–9959 - PubMed
-
- Aguilar P.S., Fröhlich F., Rehman M., Shales M., Ulitsky I., Olivera-Couto A., Braberg H., Shamir R., Walter P., Mann M., et al. 2010. A plasma-membrane E-MAP reveals links of the eisosome with sphingolipid metabolism and endosomal trafficking. Nat. Struct. Mol. Biol. 17:901–908 10.1038/nsmb.1829 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

