The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis
- PMID: 21954460
- PMCID: PMC3203420
- DOI: 10.1105/tpc.111.089870
The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis
Abstract
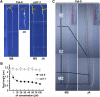
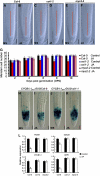
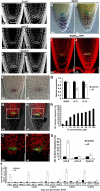
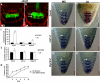
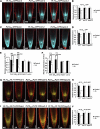
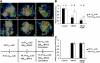
The root stem cell niche, which in the Arabidopsis thaliana root meristem is an area of four mitotically inactive quiescent cells (QCs) and the surrounding mitotically active stem cells, is critical for root development and growth. We report here that during jasmonate-induced inhibition of primary root growth, jasmonate reduces root meristem activity and leads to irregular QC division and columella stem cell differentiation. Consistently, jasmonate reduces the expression levels of the AP2-domain transcription factors PLETHORA1 (PLT1) and PLT2, which form a developmentally instructive protein gradient and mediate auxin-induced regulation of stem cell niche maintenance. Not surprisingly, the effects of jasmonate on root stem cell niche maintenance and PLT expression require the functioning of MYC2/JASMONATE INSENSITIVE1, a basic helix-loop-helix transcription factor that involves versatile aspects of jasmonate-regulated gene expression. Gel shift and chromatin immunoprecipitation experiments reveal that MYC2 directly binds the promoters of PLT1 and PLT2 and represses their expression. We propose that MYC2-mediated repression of PLT expression integrates jasmonate action into the auxin pathway in regulating root meristem activity and stem cell niche maintenance. This study illustrates a molecular framework for jasmonate-induced inhibition of root growth through interaction with the growth regulator auxin.
Figures


Similar articles
-
Jasmonate-regulated root growth inhibition and root hair elongation.J Exp Bot. 2023 Feb 13;74(4):1176-1185. doi: 10.1093/jxb/erac441. J Exp Bot. 2023. PMID: 36346644 Free PMC article. Review.
-
TIME FOR COFFEE controls root meristem size by changes in auxin accumulation in Arabidopsis.J Exp Bot. 2014 Jan;65(1):275-86. doi: 10.1093/jxb/ert374. Epub 2013 Nov 25. J Exp Bot. 2014. PMID: 24277277 Free PMC article.
-
Arabidopsis Tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche.Plant Cell. 2010 Nov;22(11):3692-709. doi: 10.1105/tpc.110.075721. Epub 2010 Nov 2. Plant Cell. 2010. PMID: 21045165 Free PMC article.
-
The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors.Plant Cell. 2012 Jul;24(7):2898-916. doi: 10.1105/tpc.112.098277. Epub 2012 Jul 20. Plant Cell. 2012. PMID: 22822206 Free PMC article.
-
The plant Mediator complex and its role in jasmonate signaling.J Exp Bot. 2019 Jul 5;70(13):3415-3424. doi: 10.1093/jxb/erz233. J Exp Bot. 2019. PMID: 31089685 Free PMC article. Review.
Cited by
-
The Physcomitrella patens unique alpha-dioxygenase participates in both developmental processes and defense responses.BMC Plant Biol. 2015 Feb 12;15:45. doi: 10.1186/s12870-015-0439-z. BMC Plant Biol. 2015. PMID: 25848849 Free PMC article.
-
Jasmonate-regulated root growth inhibition and root hair elongation.J Exp Bot. 2023 Feb 13;74(4):1176-1185. doi: 10.1093/jxb/erac441. J Exp Bot. 2023. PMID: 36346644 Free PMC article. Review.
-
AtOPR3 specifically inhibits primary root growth in Arabidopsis under phosphate deficiency.Sci Rep. 2016 Apr 22;6:24778. doi: 10.1038/srep24778. Sci Rep. 2016. PMID: 27101793 Free PMC article.
-
A complex systems approach to Arabidopsis root stem-cell niche developmental mechanisms: from molecules, to networks, to morphogenesis.Plant Mol Biol. 2012 Nov;80(4-5):351-63. doi: 10.1007/s11103-012-9954-6. Epub 2012 Sep 4. Plant Mol Biol. 2012. PMID: 22945341 Review.
-
A Jasmonate-Activated MYC2-Dof2.1-MYC2 Transcriptional Loop Promotes Leaf Senescence in Arabidopsis.Plant Cell. 2020 Jan;32(1):242-262. doi: 10.1105/tpc.19.00297. Epub 2019 Oct 22. Plant Cell. 2020. PMID: 31641025 Free PMC article.
References
-
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 - PubMed
-
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 - PubMed
-
- Alonso J.M., et al. (2003b). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Bechtold N., Pelletier G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259–266 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

