Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells
- PMID: 21939504
- PMCID: PMC3190352
- DOI: 10.1186/1476-4598-10-117
Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells
Abstract
Background: Tumor-associated macrophages (TAMs) are alternatively activated cells induced by interleukin-4 (IL-4)-releasing CD4+ T cells. TAMs promote breast cancer invasion and metastasis; however, the mechanisms underlying these interactions between macrophages and tumor cells that lead to cancer metastasis remain elusive. Previous studies have found microRNAs (miRNAs) circulating in the peripheral blood and have identified microvesicles, or exosomes, as mediators of cell-cell communication. Therefore, one alternative mechanism for the promotion of breast cancer cell invasion by TAMs may be through macrophage-secreted exosomes, which would deliver invasion-potentiating miRNAs to breast cancer cells.
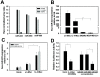
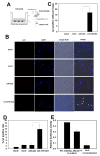
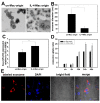
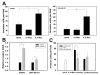
Results: We utilized a co-culture system with IL-4-activated macrophages and breast cancer cells to verify that miRNAs are transported from macrophages to breast cancer cells. The shuttling of fluorescently-labeled exogenous miRNAs from IL-4-activated macrophages to co-cultivated breast cancer cells without direct cell-cell contact was observed. miR-223, a miRNA specific for IL-4-activated macrophages, was detected within the exosomes released by macrophages and was significantly elevated in the co-cultivated SKBR3 and MDA-MB-231 cells. The invasiveness of the co-cultivated breast cancer cells decreased when the IL-4-activated macrophages were treated with a miR-223 antisense oligonucleotide (ASO) that would inhibit miR-223 expression. Furthermore, results from a functional assay revealed that miR-223 promoted the invasion of breast cancer cells via the Mef2c-β-catenin pathway.
Conclusions: We conclude that macrophages regulate the invasiveness of breast cancer cells through exosome-mediated delivery of oncogenic miRNAs. Our data provide insight into the mechanisms underlying the metastasis-promoting interactions between macrophages and breast cancer cells.
Figures
Similar articles
-
Mouse 4T1 Breast Cancer Cell-Derived Exosomes Induce Proinflammatory Cytokine Production in Macrophages via miR-183.J Immunol. 2020 Nov 15;205(10):2916-2925. doi: 10.4049/jimmunol.1901104. Epub 2020 Sep 28. J Immunol. 2020. PMID: 32989094
-
MiR-124 targets Slug to regulate epithelial-mesenchymal transition and metastasis of breast cancer.Carcinogenesis. 2013 Mar;34(3):713-22. doi: 10.1093/carcin/bgs383. Epub 2012 Dec 17. Carcinogenesis. 2013. PMID: 23250910 Free PMC article.
-
Exosome-mediated miR-33 transfer induces M1 polarization in mouse macrophages and exerts antitumor effect in 4T1 breast cancer cell line.Int Immunopharmacol. 2021 Jan;90:107198. doi: 10.1016/j.intimp.2020.107198. Epub 2020 Nov 25. Int Immunopharmacol. 2021. PMID: 33249048
-
Tumor-Associated Macrophages as Multifaceted Regulators of Breast Tumor Growth.Int J Mol Sci. 2021 Jun 18;22(12):6526. doi: 10.3390/ijms22126526. Int J Mol Sci. 2021. PMID: 34207035 Free PMC article. Review.
-
Lipid-based carriers of microRNAs and intercellular communication.Curr Opin Lipidol. 2012 Apr;23(2):91-7. doi: 10.1097/MOL.0b013e328350a425. Curr Opin Lipidol. 2012. PMID: 22418571 Free PMC article. Review.
Cited by
-
The Biology of Exosomes in Breast Cancer Progression: Dissemination, Immune Evasion and Metastatic Colonization.Cancers (Basel). 2020 Aug 5;12(8):2179. doi: 10.3390/cancers12082179. Cancers (Basel). 2020. PMID: 32764376 Free PMC article. Review.
-
Reciprocal interactions between endothelial cells and macrophages in angiogenic vascular niches.Exp Cell Res. 2013 Jul 1;319(11):1626-34. doi: 10.1016/j.yexcr.2013.03.026. Epub 2013 Mar 28. Exp Cell Res. 2013. PMID: 23542777 Free PMC article. Review.
-
Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes.Sci Rep. 2015 Jun 3;5:10300. doi: 10.1038/srep10300. Sci Rep. 2015. PMID: 26036864 Free PMC article.
-
MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment.Oncogene. 2015 Nov 26;34(48):5857-68. doi: 10.1038/onc.2015.89. Epub 2015 Apr 13. Oncogene. 2015. PMID: 25867073 Free PMC article. Review.
-
Aspects of the Tumor Microenvironment Involved in Immune Resistance and Drug Resistance.Front Immunol. 2021 May 27;12:656364. doi: 10.3389/fimmu.2021.656364. eCollection 2021. Front Immunol. 2021. PMID: 34122412 Free PMC article. Review.
References
-
- Leek RD, Lewis CE, Whitehouse R, Greenall M, Clarke J, Harris AL. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res. 1996;56:4625–4629. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

