Post-translational modifications and lipid binding profile of insect cell-expressed full-length mammalian synaptotagmin 1
- PMID: 21928778
- PMCID: PMC3217305
- DOI: 10.1021/bi200998y
Post-translational modifications and lipid binding profile of insect cell-expressed full-length mammalian synaptotagmin 1
Abstract
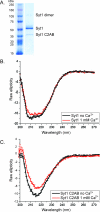
Synaptotagmin 1 (Syt1) is a Ca(2+) sensor for SNARE-mediated, Ca(2+)-triggered synaptic vesicle fusion in neurons. It is composed of luminal, transmembrane, linker, and two Ca(2+)-binding (C2) domains. Here we describe expression and purification of full-length mammalian Syt1 in insect cells along with an extensive biochemical characterization of the purified protein. The expressed and purified protein is properly folded and has increased α-helical content compared to the C2AB fragment alone. Post-translational modifications of Syt1 were analyzed by mass spectrometry, revealing the same modifications of Syt1 that were previously described for Syt1 purified from brain extract or mammalian cell lines, along with a novel modification of Syt1, tyrosine nitration. A lipid binding screen with both full-length Syt1 and the C2AB fragments of Syt1 and Syt3 isoforms revealed new Syt1-lipid interactions. These results suggest a conserved lipid binding mechanism in which Ca(2+)-independent interactions are mediated via a lysine rich region of the C2B domain while Ca(2+)-dependent interactions are mediated via the Ca(2+)-binding loops.
© 2011 American Chemical Society
Figures

Similar articles
-
The juxtamembrane linker of full-length synaptotagmin 1 controls oligomerization and calcium-dependent membrane binding.J Biol Chem. 2014 Aug 8;289(32):22161-71. doi: 10.1074/jbc.M114.569327. Epub 2014 Jun 27. J Biol Chem. 2014. PMID: 24973220 Free PMC article.
-
A Post-Docking Role of Synaptotagmin 1-C2B Domain Bottom Residues R398/399 in Mouse Chromaffin Cells.J Neurosci. 2015 Oct 21;35(42):14172-82. doi: 10.1523/JNEUROSCI.1911-15.2015. J Neurosci. 2015. PMID: 26490858 Free PMC article.
-
SNARE complex alters the interactions of the Ca2+ sensor synaptotagmin 1 with lipid bilayers.Biophys J. 2021 Feb 16;120(4):642-661. doi: 10.1016/j.bpj.2020.12.025. Epub 2021 Jan 14. Biophys J. 2021. PMID: 33453271 Free PMC article.
-
Models of synaptotagmin-1 to trigger Ca2+ -dependent vesicle fusion.FEBS Lett. 2018 Nov;592(21):3480-3492. doi: 10.1002/1873-3468.13193. Epub 2018 Jul 30. FEBS Lett. 2018. PMID: 30004579 Review.
-
SYT1-Associated Neurodevelopmental Disorder: A Narrative Review.Children (Basel). 2022 Sep 22;9(10):1439. doi: 10.3390/children9101439. Children (Basel). 2022. PMID: 36291375 Free PMC article. Review.
Cited by
-
Controlling synaptotagmin activity by electrostatic screening.Nat Struct Mol Biol. 2012 Oct;19(10):991-7. doi: 10.1038/nsmb.2375. Epub 2012 Sep 2. Nat Struct Mol Biol. 2012. PMID: 22940675 Free PMC article.
-
Calcium Sensors of Neurotransmitter Release.Adv Neurobiol. 2023;33:119-138. doi: 10.1007/978-3-031-34229-5_5. Adv Neurobiol. 2023. PMID: 37615865
-
Vesicle trafficking and vesicle fusion: mechanisms, biological functions, and their implications for potential disease therapy.Mol Biomed. 2022 Sep 21;3(1):29. doi: 10.1186/s43556-022-00090-3. Mol Biomed. 2022. PMID: 36129576 Free PMC article. Review.
-
Gangliosides interact with synaptotagmin to form the high-affinity receptor complex for botulinum neurotoxin B.Proc Natl Acad Sci U S A. 2019 Sep 3;116(36):18098-18108. doi: 10.1073/pnas.1908051116. Epub 2019 Aug 20. Proc Natl Acad Sci U S A. 2019. PMID: 31431523 Free PMC article.
-
Hydrophobic contributions to the membrane docking of synaptotagmin 7 C2A domain: mechanistic contrast between isoforms 1 and 7.Biochemistry. 2012 Oct 2;51(39):7654-64. doi: 10.1021/bi3007115. Epub 2012 Sep 21. Biochemistry. 2012. PMID: 22966849 Free PMC article.
References
-
- Perin M. S.; Brose N.; Jahn R.; Sudhof T. C. (1991) Domain structure of synaptotagmin (p65). J. Biol. Chem. 266, 623–629. - PubMed
-
- Han W.; Rhee J. S.; Maximov A.; Lao Y.; Mashimo T.; Rosenmund C.; Sudhof T. C. (2004) N-Glycosylation is essential for vesicular targeting of synaptotagmin 1. Neuron 41, 85–99. - PubMed
-
- Fukuda M. (2002) Vesicle-associated membrane protein-2/synaptobrevin binding to synaptotagmin I promotes O-glycosylation of synaptotagmin I. J. Biol. Chem. 277, 30351–30358. - PubMed
-
- Kanno E.; Fukuda M. (2008) Increased plasma membrane localization of O-glycosylation-deficient mutant of synaptotagmin I in PC12 cells. J. Neurosci. Res. 86, 1036–1043. - PubMed
-
- Veit M.; Sollner T. H.; Rothman J. E. (1996) Multiple palmitoylation of synaptotagmin and the t-SNARE SNAP-25. FEBS Lett. 385, 119–123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

