Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles
- PMID: 21921995
- PMCID: PMC3171997
- DOI: 10.1021/ct200162x
Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles
Abstract
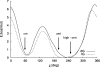
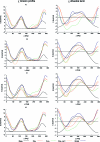
We report a reparameterization of the glycosidic torsion χ of the Cornell et al. AMBER force field for RNA, χ(OL). The parameters remove destabilization of the anti region found in the ff99 force field and thus prevent formation of spurious ladder-like structural distortions in RNA simulations. They also improve the description of the syn region and the syn-anti balance as well as enhance MD simulations of various RNA structures. Although χ(OL) can be combined with both ff99 and ff99bsc0, we recommend the latter. We do not recommend using χ(OL) for B-DNA because it does not improve upon ff99bsc0 for canonical structures. However, it might be useful in simulations of DNA molecules containing syn nucleotides. Our parametrization is based on high-level QM calculations and differs from conventional parametrization approaches in that it incorporates some previously neglected solvation-related effects (which appear to be essential for obtaining correct anti/high-anti balance). Our χ(OL) force field is compared with several previous glycosidic torsion parametrizations.
Figures
Similar articles
-
Performance of Molecular Mechanics Force Fields for RNA Simulations: Stability of UUCG and GNRA Hairpins.J Chem Theory Comput. 2010 Dec 14;6(12):3836-3849. doi: 10.1021/ct100481h. Epub 2010 Nov 9. J Chem Theory Comput. 2010. PMID: 35283696 Free PMC article.
-
Molecular dynamics and quantum mechanics of RNA: conformational and chemical change we can believe in.Acc Chem Res. 2010 Jan 19;43(1):40-7. doi: 10.1021/ar900093g. Acc Chem Res. 2010. PMID: 19754142 Free PMC article.
-
Toward Improved Description of DNA Backbone: Revisiting Epsilon and Zeta Torsion Force Field Parameters.J Chem Theory Comput. 2013 May 14;9(5):2339-2354. doi: 10.1021/ct400154j. J Chem Theory Comput. 2013. PMID: 24058302 Free PMC article.
-
Molecular dynamic simulations of environment and sequence dependent DNA conformations: the development of the BMS nucleic acid force field and comparison with experimental results.J Biomol Struct Dyn. 1998 Dec;16(3):487-509. doi: 10.1080/07391102.1998.10508265. J Biomol Struct Dyn. 1998. PMID: 10052609 Review.
-
A perspective on the primary and three-dimensional structures of carbohydrates.Carbohydr Res. 2013 Aug 30;378:123-32. doi: 10.1016/j.carres.2013.02.005. Epub 2013 Feb 24. Carbohydr Res. 2013. PMID: 23522728 Review.
Cited by
-
A general RNA force field: comprehensive analysis of energy minima of molecular fragments of RNA.J Mol Model. 2021 Apr 26;27(5):137. doi: 10.1007/s00894-021-04746-9. J Mol Model. 2021. PMID: 33903935
-
Molecular Simulations Matching Denaturation Experiments for N6-Methyladenosine.ACS Cent Sci. 2022 Aug 24;8(8):1218-1228. doi: 10.1021/acscentsci.2c00565. Epub 2022 Aug 3. ACS Cent Sci. 2022. PMID: 36032773 Free PMC article.
-
Relative stability of different DNA guanine quadruplex stem topologies derived using large-scale quantum-chemical computations.J Am Chem Soc. 2013 Jul 3;135(26):9785-96. doi: 10.1021/ja402525c. Epub 2013 Jun 19. J Am Chem Soc. 2013. PMID: 23742743 Free PMC article.
-
Molecular simulations of RNA 2'-O-transesterification reaction models in solution.J Phys Chem B. 2013 Jan 10;117(1):94-103. doi: 10.1021/jp3084277. Epub 2012 Dec 24. J Phys Chem B. 2013. PMID: 23214417 Free PMC article.
-
Binding of the peptide deformylase on the ribosome surface modulates the exit tunnel interior.Biophys J. 2022 Dec 6;121(23):4443-4451. doi: 10.1016/j.bpj.2022.11.004. Epub 2022 Nov 5. Biophys J. 2022. PMID: 36335428 Free PMC article.
References
-
- Weiner S. J.; Kollman P. A.; Case D. A.; Singh U. C.; Ghio C.; Alagona G.; Profeta S.; Weiner P. J. Am. Chem. Soc. 1984, 106 (3), 765–784.
-
- Weiner S. J.; Kollman P. A.; Nguyen D. T.; Case D. A. J. Comput. Chem. 1986, 7 (2), 230–252. - PubMed
-
- Mackerell A. D. J. Comput. Chem. 2004, 25 (13), 1584–1604. - PubMed
-
- Orozco M.; Noy A.; Perez A. Curr. Opin. Struct. Biol. 2008, 18 (2), 185–193. - PubMed
-
- Fulle S.; Gohlke H. J. Mol. Recognit. 2010, 23 (2), 220–231. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
