Structural analysis of macromolecular assemblies by electron microscopy
- PMID: 21919528
- PMCID: PMC3239172
- DOI: 10.1021/cr100353t
Structural analysis of macromolecular assemblies by electron microscopy
Figures


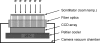

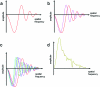
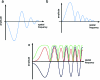





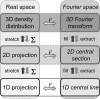






Similar articles
-
Metal shadowing and decoration in electron microscopy of biological macromolecules.Ann N Y Acad Sci. 1986;483:57-76. doi: 10.1111/j.1749-6632.1986.tb34497.x. Ann N Y Acad Sci. 1986. PMID: 2436521 No abstract available.
-
Determining the structure of biological macromolecules by transmission electron microscopy, single particle analysis and 3D reconstruction.Prog Biophys Mol Biol. 2001;75(3):121-64. doi: 10.1016/s0079-6107(01)00004-9. Prog Biophys Mol Biol. 2001. PMID: 11376797 Review.
-
Survey of the analysis of continuous conformational variability of biological macromolecules by electron microscopy.Acta Crystallogr F Struct Biol Commun. 2019 Jan 1;75(Pt 1):19-32. doi: 10.1107/S2053230X18015108. Epub 2019 Jan 1. Acta Crystallogr F Struct Biol Commun. 2019. PMID: 30605122 Free PMC article. Review.
-
Low temperature electron microscopy.Annu Rev Biophys Bioeng. 1981;10:133-49. doi: 10.1146/annurev.bb.10.060181.001025. Annu Rev Biophys Bioeng. 1981. PMID: 7020572 Review. No abstract available.
-
Structural studies by electron tomography: from cells to molecules.Annu Rev Biochem. 2005;74:833-65. doi: 10.1146/annurev.biochem.73.011303.074112. Annu Rev Biochem. 2005. PMID: 15952904 Review.
Cited by
-
(Cryo)Transmission Electron Microscopy of Phospholipid Model Membranes Interacting with Amphiphilic and Polyphilic Molecules.Polymers (Basel). 2017 Oct 19;9(10):521. doi: 10.3390/polym9100521. Polymers (Basel). 2017. PMID: 30965829 Free PMC article. Review.
-
Membranes under the Magnetic Lens: A Dive into the Diverse World of Membrane Protein Structures Using Cryo-EM.Chem Rev. 2022 Sep 14;122(17):13989-14017. doi: 10.1021/acs.chemrev.1c00837. Epub 2022 Jul 18. Chem Rev. 2022. PMID: 35849490 Free PMC article. Review.
-
Isotropic reconstruction for electron tomography with deep learning.Nat Commun. 2022 Oct 29;13(1):6482. doi: 10.1038/s41467-022-33957-8. Nat Commun. 2022. PMID: 36309499 Free PMC article.
-
Assembly of macromolecular complexes by satisfaction of spatial restraints from electron microscopy images.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18821-6. doi: 10.1073/pnas.1216549109. Epub 2012 Oct 29. Proc Natl Acad Sci U S A. 2012. PMID: 23112201 Free PMC article.
-
Cryogenic superresolution correlative light and electron microscopy on the frontier of subcellular imaging.Biophys Rev. 2021 Nov 26;13(6):1163-1171. doi: 10.1007/s12551-021-00851-4. eCollection 2021 Dec. Biophys Rev. 2021. PMID: 35059034 Free PMC article. Review.
References
-
- Dubochet J.; Adrian M.; Chang J. J.; Homo J. C.; Lepault J.; McDowall A. W.; Schultz P. Q. Rev. Biophys. 1988, 21, 129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

