A physical interaction network of dengue virus and human proteins
- PMID: 21911577
- PMCID: PMC3237087
- DOI: 10.1074/mcp.M111.012187
A physical interaction network of dengue virus and human proteins
Abstract
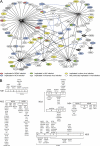
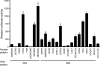
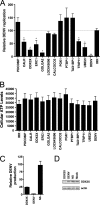
Dengue virus (DENV), an emerging mosquito-transmitted pathogen capable of causing severe disease in humans, interacts with host cell factors to create a more favorable environment for replication. However, few interactions between DENV and human proteins have been reported to date. To identify DENV-human protein interactions, we used high-throughput yeast two-hybrid assays to screen the 10 DENV proteins against a human liver activation domain library. From 45 DNA-binding domain clones containing either full-length viral genes or partially overlapping gene fragments, we identified 139 interactions between DENV and human proteins, the vast majority of which are novel. These interactions involved 105 human proteins, including six previously implicated in DENV infection and 45 linked to the replication of other viruses. Human proteins with functions related to the complement and coagulation cascade, the centrosome, and the cytoskeleton were enriched among the DENV interaction partners. To determine if the cellular proteins were required for DENV infection, we used small interfering RNAs to inhibit their expression. Six of 12 proteins targeted (CALR, DDX3X, ERC1, GOLGA2, TRIP11, and UBE2I) caused a significant decrease in the replication of a DENV replicon. We further showed that calreticulin colocalized with viral dsRNA and with the viral NS3 and NS5 proteins in DENV-infected cells, consistent with a direct role for calreticulin in DENV replication. Human proteins that interacted with DENV had significantly higher average degree and betweenness than expected by chance, which provides additional support for the hypothesis that viruses preferentially target cellular proteins that occupy central position in the human protein interaction network. This study provides a valuable starting point for additional investigations into the roles of human proteins in DENV infection.
Figures
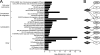
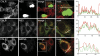
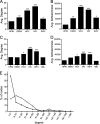
Similar articles
-
SUMO Modification Stabilizes Dengue Virus Nonstructural Protein 5 To Support Virus Replication.J Virol. 2016 Apr 14;90(9):4308-4319. doi: 10.1128/JVI.00223-16. Print 2016 May. J Virol. 2016. PMID: 26889037 Free PMC article.
-
Caveolin-1 in lipid rafts interacts with dengue virus NS3 during polyprotein processing and replication in HMEC-1 cells.PLoS One. 2014 Mar 18;9(3):e90704. doi: 10.1371/journal.pone.0090704. eCollection 2014. PLoS One. 2014. PMID: 24643062 Free PMC article.
-
Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication.J Virol. 2014 Jun;88(12):6793-804. doi: 10.1128/JVI.00045-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696471 Free PMC article.
-
The Transactions of NS3 and NS5 in Flaviviral RNA Replication.Adv Exp Med Biol. 2018;1062:147-163. doi: 10.1007/978-981-10-8727-1_11. Adv Exp Med Biol. 2018. PMID: 29845531 Review.
-
Early dengue virus interactions: the role of dendritic cells during infection.Virus Res. 2016 Sep 2;223:88-98. doi: 10.1016/j.virusres.2016.07.001. Epub 2016 Jul 2. Virus Res. 2016. PMID: 27381061 Review.
Cited by
-
A binary interaction map between turnip mosaic virus and Arabidopsis thaliana proteomes.Commun Biol. 2023 Jan 11;6(1):28. doi: 10.1038/s42003-023-04427-8. Commun Biol. 2023. PMID: 36631662 Free PMC article.
-
Exploring the Implication of DDX3X in DENV Infection: Discovery of the First-in-Class DDX3X Fluorescent Inhibitor.ACS Med Chem Lett. 2020 Apr 9;11(5):956-962. doi: 10.1021/acsmedchemlett.9b00681. eCollection 2020 May 14. ACS Med Chem Lett. 2020. PMID: 32435411 Free PMC article.
-
Oxidative stress influences positive strand RNA virus genome synthesis and capping.Virology. 2015 Jan 15;475:219-29. doi: 10.1016/j.virol.2014.10.037. Epub 2014 Dec 13. Virology. 2015. PMID: 25514423 Free PMC article.
-
DEAD-Box Helicase DDX25 Is a Negative Regulator of Type I Interferon Pathway and Facilitates RNA Virus Infection.Front Cell Infect Microbiol. 2017 Aug 4;7:356. doi: 10.3389/fcimb.2017.00356. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28824886 Free PMC article.
-
Akt Interacts with Usutu Virus Polymerase, and Its Activity Modulates Viral Replication.Pathogens. 2021 Feb 20;10(2):244. doi: 10.3390/pathogens10020244. Pathogens. 2021. PMID: 33672588 Free PMC article.
References
-
- Llano M., Saenz D. T., Meehan A., Wongthida P., Peretz M., Walker W. H., Teo W., Poeschla E. M. (2006) An essential role for LEDGF/p75 in HIV integration. Science 314, 461–464 - PubMed
-
- Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y. (2003) LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278, 33528–33539 - PubMed
-
- Galluzzi L., Kepp O., Morselli E., Vitale I., Senovilla L., Pinti M., Zitvogel L., Kroemer G. (2010) Viral strategies for the evasion of immunogenic cell death. J. Intern Med. 267, 526–542 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

