The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO
- PMID: 21909281
- PMCID: PMC3164695
- DOI: 10.1371/journal.pgen.1002235
The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO
Abstract
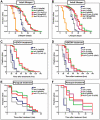
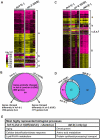
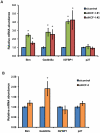
The conserved DAF-16/FOXO transcription factors and SIR-2.1/SIRT1 deacetylases are critical for diverse biological processes, particularly longevity and stress response; and complex regulation of DAF-16/FOXO by SIR-2.1/SIRT1 is central to appropriate biological outcomes. Caenorhabditis elegans Host Cell Factor 1 (HCF-1) is a longevity determinant previously shown to act as a co-repressor of DAF-16. We report here that HCF-1 represents an integral player in the regulatory loop linking SIR-2.1/SIRT1 and DAF-16/FOXO in both worms and mammals. Genetic analyses showed that hcf-1 acts downstream of sir-2.1 to influence lifespan and oxidative stress response in C. elegans. Gene expression profiling revealed a striking 80% overlap between the DAF-16 target genes responsive to hcf-1 mutation and sir-2.1 overexpression. Subsequent GO-term analyses of HCF-1 and SIR-2.1-coregulated DAF-16 targets suggested that HCF-1 and SIR-2.1 together regulate specific aspects of DAF-16-mediated transcription particularly important for aging and stress responses. Analogous to its role in regulating DAF-16/SIR-2.1 target genes in C. elegans, the mammalian HCF-1 also repressed the expression of several FOXO/SIRT1 target genes. Protein-protein association studies demonstrated that SIR-2.1/SIRT1 and HCF-1 form protein complexes in worms and mammalian cells, highlighting the conservation of their regulatory relationship. Our findings uncover a conserved interaction between the key longevity determinants SIR-2.1/SIRT1 and HCF-1, and they provide new insights into the complex regulation of FOXO proteins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


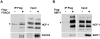
Similar articles
-
Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator.PLoS Biol. 2008 Sep 30;6(9):e233. doi: 10.1371/journal.pbio.0060233. PLoS Biol. 2008. PMID: 18828672 Free PMC article.
-
Royal Jelly-Mediated Prolongevity and Stress Resistance in Caenorhabditis elegans Is Possibly Modulated by the Interplays of DAF-16, SIR-2.1, HCF-1, and 14-3-3 Proteins.J Gerontol A Biol Sci Med Sci. 2015 Jul;70(7):827-38. doi: 10.1093/gerona/glu120. Epub 2014 Jul 29. J Gerontol A Biol Sci Med Sci. 2015. PMID: 25073462
-
Host cell factor 1 inhibits SKN-1 to modulate oxidative stress responses in Caenorhabditis elegans.Aging Cell. 2012 Aug;11(4):717-21. doi: 10.1111/j.1474-9726.2012.00831.x. Epub 2012 Jun 4. Aging Cell. 2012. PMID: 22568582 Free PMC article.
-
A stress response pathway involving sirtuins, forkheads and 14-3-3 proteins.Cell Cycle. 2006 Nov;5(22):2588-91. doi: 10.4161/cc.5.22.3513. Epub 2006 Nov 15. Cell Cycle. 2006. PMID: 17172829 Review.
-
The search for DAF-16/FOXO transcriptional targets: approaches and discoveries.Exp Gerontol. 2006 Oct;41(10):910-21. doi: 10.1016/j.exger.2006.06.040. Epub 2006 Aug 24. Exp Gerontol. 2006. PMID: 16934425 Review.
Cited by
-
Elevated Trehalose Levels in C. elegans daf-2 Mutants Increase Stress Resistance, Not Lifespan.Metabolites. 2021 Feb 12;11(2):105. doi: 10.3390/metabo11020105. Metabolites. 2021. PMID: 33673074 Free PMC article.
-
Yeast replicative aging: a paradigm for defining conserved longevity interventions.FEMS Yeast Res. 2014 Feb;14(1):148-59. doi: 10.1111/1567-1364.12104. Epub 2013 Oct 30. FEMS Yeast Res. 2014. PMID: 24119093 Free PMC article. Review.
-
The sirtuin SIRT6 regulates lifespan in male mice.Nature. 2012 Feb 22;483(7388):218-21. doi: 10.1038/nature10815. Nature. 2012. PMID: 22367546
-
Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide.Nat Chem Biol. 2013 Nov;9(11):693-700. doi: 10.1038/nchembio.1352. Epub 2013 Sep 29. Nat Chem Biol. 2013. PMID: 24077178 Free PMC article.
-
Ageing: longevity hits a roadblock.Nature. 2011 Sep 21;477(7365):410-1. doi: 10.1038/477410a. Nature. 2011. PMID: 21938058 No abstract available.
References
-
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. - PubMed
-
- Burgering BM, Kops GJ. Cell cycle and death control: long live Forkheads. Trends Biochem Sci. 2002;27:352–360. - PubMed
-
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. - PubMed
-
- Giannakou ME, Goss M, Junger MA, Hafen E, Leevers SJ, et al. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. - PubMed
-
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

