Genome-wide comparison of African-ancestry populations from CARe and other cohorts reveals signals of natural selection
- PMID: 21907010
- PMCID: PMC3169818
- DOI: 10.1016/j.ajhg.2011.07.025
Genome-wide comparison of African-ancestry populations from CARe and other cohorts reveals signals of natural selection
Abstract
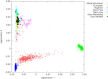
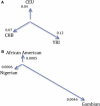
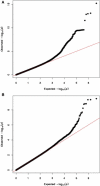
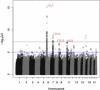
The study of recent natural selection in human populations has important applications to human history and medicine. Positive natural selection drives the increase in beneficial alleles and plays a role in explaining diversity across human populations. By discovering traits subject to positive selection, we can better understand the population level response to environmental pressures including infectious disease. Our study examines unusual population differentiation between three large data sets to detect natural selection. The populations examined, African Americans, Nigerians, and Gambians, are genetically close to one another (F(ST) < 0.01 for all pairs), allowing us to detect selection even with moderate changes in allele frequency. We also develop a tree-based method to pinpoint the population in which selection occurred, incorporating information across populations. Our genome-wide significant results corroborate loci previously reported to be under selection in Africans including HBB and CD36. At the HLA locus on chromosome 6, results suggest the existence of multiple, independent targets of population-specific selective pressure. In addition, we report a genome-wide significant (p = 1.36 × 10(-11)) signal of selection in the prostate stem cell antigen (PSCA) gene. The most significantly differentiated marker in our analysis, rs2920283, is highly differentiated in both Africa and East Asia and has prior genome-wide significant associations to bladder and gastric cancers.
Copyright © 2011 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Genome-wide scan of 29,141 African Americans finds no evidence of directional selection since admixture.Am J Hum Genet. 2014 Oct 2;95(4):437-44. doi: 10.1016/j.ajhg.2014.08.011. Epub 2014 Sep 18. Am J Hum Genet. 2014. PMID: 25242497 Free PMC article.
-
Spatial and temporal diversity of positive selection on shared haplotypes at the PSCA locus among worldwide human populations.Heredity (Edinb). 2023 Aug;131(2):156-169. doi: 10.1038/s41437-023-00631-8. Epub 2023 Jun 23. Heredity (Edinb). 2023. PMID: 37353592 Free PMC article.
-
Genome-wide detection of natural selection in African Americans pre- and post-admixture.Genome Res. 2012 Mar;22(3):519-27. doi: 10.1101/gr.124784.111. Epub 2011 Nov 29. Genome Res. 2012. PMID: 22128132 Free PMC article.
-
The genomic landscape of African populations in health and disease.Hum Mol Genet. 2017 Oct 1;26(R2):R225-R236. doi: 10.1093/hmg/ddx253. Hum Mol Genet. 2017. PMID: 28977439 Free PMC article. Review.
-
Genetic variation and adaptation in Africa: implications for human evolution and disease.Cold Spring Harb Perspect Biol. 2014 Jul 1;6(7):a008524. doi: 10.1101/cshperspect.a008524. Cold Spring Harb Perspect Biol. 2014. PMID: 24984772 Free PMC article. Review.
Cited by
-
Convergent evolution in European and Rroma populations reveals pressure exerted by plague on Toll-like receptors.Proc Natl Acad Sci U S A. 2014 Feb 18;111(7):2668-73. doi: 10.1073/pnas.1317723111. Epub 2014 Feb 3. Proc Natl Acad Sci U S A. 2014. PMID: 24550294 Free PMC article.
-
Limited evidence for classic selective sweeps in African populations.Genetics. 2012 Nov;192(3):1049-64. doi: 10.1534/genetics.112.144071. Epub 2012 Sep 7. Genetics. 2012. PMID: 22960214 Free PMC article.
-
How HLA diversity is apportioned: influence of selection and relevance to transplantation.Philos Trans R Soc Lond B Biol Sci. 2022 Jun 6;377(1852):20200420. doi: 10.1098/rstb.2020.0420. Epub 2022 Apr 18. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35430892 Free PMC article.
-
Population genomics of wild Chinese rhesus macaques reveals a dynamic demographic history and local adaptation, with implications for biomedical research.Gigascience. 2018 Sep 1;7(9):giy106. doi: 10.1093/gigascience/giy106. Gigascience. 2018. PMID: 30165519 Free PMC article.
-
The Effect of Balancing Selection on Population Differentiation: A Study with HLA Genes.G3 (Bethesda). 2018 Jul 31;8(8):2805-2815. doi: 10.1534/g3.118.200367. G3 (Bethesda). 2018. PMID: 29950428 Free PMC article.
References
-
- Sabeti P.C., Reich D.E., Higgins J.M., Levine H.Z., Richter D.J., Schaffner S.F., Gabriel S.B., Platko J.V., Patterson N.J., McDonald G.J. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. - PubMed
-
- Ko W.Y., Kaercher K.A., Giombini E., Marcatili P., Froment A., Ibrahim M., Lema G., Nyambo T.B., Omar S.A., Wambebe C. Effects of natural selection and gene conversion on the evolution of human glycophorins coding for MNS blood polymorphisms in malaria-endemic African populations. Am. J. Hum. Genet. 2011;88:741–754. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

