Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases
- PMID: 21885342
- PMCID: PMC3225699
- DOI: 10.1016/j.molmed.2011.07.005
Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases
Abstract
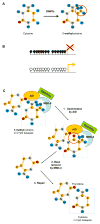
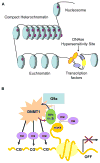
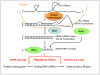
The pathogenic origin of autoimmune diseases can be traced to both genetic susceptibility and epigenetic modifications arising from exposure to the environment. Epigenetic modifications influence gene expression and alter cellular functions without modifying the genomic sequence. CpG-DNA methylation, histone tail modifications and microRNAs (miRNAs) are the main epigenetic mechanisms of gene regulation. Understanding the molecular mechanisms that are involved in the pathophysiology of autoimmune diseases is essential for the introduction of effective, target-directed and tolerated therapies. In this review, we summarize recent findings that signify the importance of epigenetic modifications in autoimmune disorders while focusing on systemic lupus erythematosus. We also discuss future directions in basic research, autoimmune diagnostics and applied therapy.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors have no conflicts of interest.
Figures
Similar articles
-
Epigenetics in lupus.Autoimmunity. 2014 Jun;47(4):213-4. doi: 10.3109/08916934.2014.915393. Autoimmunity. 2014. PMID: 24826881
-
The Role of Epigenetics in Autoimmune/Inflammatory Disease.Front Immunol. 2019 Jul 4;10:1525. doi: 10.3389/fimmu.2019.01525. eCollection 2019. Front Immunol. 2019. PMID: 31333659 Free PMC article. Review.
-
Effects of Major Epigenetic Factors on Systemic Lupus Erythematosus.Iran Biomed J. 2018 Sep;22(5):294-302. doi: 10.29252/ibj.22.5.294. Epub 2018 May 27. Iran Biomed J. 2018. PMID: 29803202 Free PMC article. Review.
-
MicroRNAs in lupus.Autoimmunity. 2014 Jun;47(4):272-85. doi: 10.3109/08916934.2014.915955. Autoimmunity. 2014. PMID: 24826805 Free PMC article. Review.
-
Epigenetic perspectives in systemic lupus erythematosus: pathogenesis, biomarkers, and therapeutic potentials.Clin Rev Allergy Immunol. 2010 Aug;39(1):3-9. doi: 10.1007/s12016-009-8165-7. Clin Rev Allergy Immunol. 2010. PMID: 19639427 Review.
Cited by
-
DNA Methylation Patterns in CD8+ T Cells Discern Psoriasis From Psoriatic Arthritis and Correlate With Cutaneous Disease Activity.Front Cell Dev Biol. 2021 Oct 21;9:746145. doi: 10.3389/fcell.2021.746145. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34746142 Free PMC article.
-
[Systemic lupus erythematosus-are children small adults?].Z Rheumatol. 2022 Feb;81(1):28-35. doi: 10.1007/s00393-021-01116-x. Epub 2021 Nov 8. Z Rheumatol. 2022. PMID: 34748078 Review. German.
-
Genome-Wide DNA Methylation Profiles Reveal Common Epigenetic Patterns of Interferon-Related Genes in Multiple Autoimmune Diseases.Front Genet. 2019 Apr 5;10:223. doi: 10.3389/fgene.2019.00223. eCollection 2019. Front Genet. 2019. PMID: 31024609 Free PMC article.
-
Estradiol regulates the expression of CD45 splicing isoforms in lymphocytes.Mol Biol Rep. 2020 Apr;47(4):3025-3030. doi: 10.1007/s11033-020-05373-y. Epub 2020 Mar 13. Mol Biol Rep. 2020. PMID: 32170460
-
Sex differences in the expression of lupus-associated miRNAs in splenocytes from lupus-prone NZB/WF1 mice.Biol Sex Differ. 2013 Nov 1;4(1):19. doi: 10.1186/2042-6410-4-19. Biol Sex Differ. 2013. PMID: 24175965 Free PMC article.
References
-
- Hedrich CM. Genetic Variation and Epigenetic Patterns in Autoimmunity. J Genet Syndr Gene Ther. 2011 doi: 10.4172/2157-7412.10000e2. ( http://omicsonline.org) - DOI
-
- Renaudineau Y, Youinou P. Epigenetics and autoimmunity, with special emphasis on methylation. Keio J Med. 2011;60(1):10–6. - PubMed
-
- Gualtierotti R, et al. Updating on the pathogenesis of systemic lupus erythematosus. Autoimmun Rev. 2010;10(1):3–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

